Nucleic acid amplification strategies for DNA microarray-based pathogen detection
- PMID: 15128566
- PMCID: PMC404398
- DOI: 10.1128/AEM.70.5.3047-3054.2004
Nucleic acid amplification strategies for DNA microarray-based pathogen detection
Abstract
DNA microarray-based screening and diagnostic technologies have long promised comprehensive testing capabilities. However, the potential of these powerful tools has been limited by front-end target-specific nucleic acid amplification. Despite the sensitivity and specificity associated with PCR amplification, the inherent bias and limited throughput of this approach constrain the principal benefits of downstream microarray-based applications, especially for pathogen detection. To begin addressing alternative approaches, we investigated four front-end amplification strategies: random primed, isothermal Klenow fragment-based, phi29 DNA polymerase-based, and multiplex PCR. The utility of each amplification strategy was assessed by hybridizing amplicons to microarrays consisting of 70-mer oligonucleotide probes specific for enterohemorrhagic Escherichia coli O157:H7 and by quantitating their sensitivities for the detection of O157:H7 in laboratory and environmental samples. Although nearly identical levels of hybridization specificity were achieved for each method, multiplex PCR was at least 3 orders of magnitude more sensitive than any individual random amplification approach. However, the use of Klenow-plus-Klenow and phi29 polymerase-plus-Klenow tandem random amplification strategies provided better sensitivities than multiplex PCR. In addition, amplification biases among the five genetic loci tested were 2- to 20-fold for the random approaches, in contrast to >4 orders of magnitude for multiplex PCR. The same random amplification strategies were also able to detect all five diagnostic targets in a spiked environmental water sample that contained a 63-fold excess of contaminating DNA. The results presented here underscore the feasibility of using random amplification approaches and begin to systematically address the versatility of these approaches for unbiased pathogen detection from environmental sources.
Figures

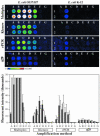
References
-
- Charles, P. T., G. J. Vora, J. D. Andreadis, A. J. Fortney, C. E. Meador, C. S. Dulcey, and D. A. Stenger. 2003. Fabrication and surface characterization of DNA microarrays using amine- and thiol-terminated oligonucleotide probes. Langmuir 19:1586-1591.
-
- Detter, J. C., J. M. Jett, S. M. Lucas, E. Dalin, A. R. Arellano, M. Wang, J. R. Nelson, J. Chapman, Y. Lou, D. Rokhsar, T. L. Hawkins, and P. M. Richardson. 2002. Isothermal strand-displacement amplification applications for high-throughput genomics. Genomics 80:691-698. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

