Gonadotropin-releasing hormone regulates expression of the DNA damage repair gene, Fanconi anemia A, in pituitary gonadotroph cells
- PMID: 15128600
- PMCID: PMC1950776
- DOI: 10.1095/biolreprod.104.030569
Gonadotropin-releasing hormone regulates expression of the DNA damage repair gene, Fanconi anemia A, in pituitary gonadotroph cells
Abstract
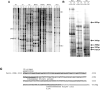
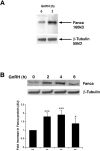
Gonadal function is critically dependant on regulated secretion of the gonadotropin hormones from anterior pituitary gonadotroph cells. Gonadotropin biosynthesis and release is triggered by the binding of hypothalamic GnRH to GnRH receptor expressed on the gonadotroph cell surface. The repertoire of regulatory molecules involved in this process are still being defined. We used the mouse L beta T2 gonadotroph cell line, which expresses both gonadotropin hormones, as a model to investigate GnRH regulation of gene expression and differential display reverse transcription-polymerase chain reaction (RT-PCR) to identify and isolate hormonally induced changes. This approach identified Fanconi anemia a (Fanca), a gene implicated in DNA damage repair, as a differentially expressed transcript. Mutations in Fanca account for the majority of cases of Fanconi anemia (FA), a recessively inherited disease identified by congenital defects, bone marrow failure, infertility, and cancer susceptibility. We confirmed expression and hormonal regulation of Fanca mRNA by quantitative RT-PCR, which showed that GnRH induced a rapid, transient increase in Fanca mRNA. Fanca protein was also acutely upregulated after GnRH treatment of L beta T2 cells. In addition, Fanca gene expression was confined to mature pituitary gonadotrophs and adult mouse pituitary and was not expressed in the immature alpha T3-1 gonadotroph cell line. Thus, this study extends the expression profile of Fanca into a highly specialized endocrine cell and demonstrates hormonal regulation of expression of the Fanca locus. We suggest that this regulatory mechanism may have a crucial role in the GnRH-response mechanism of mature gonadotrophs and perhaps the etiology of FA.
Figures



References
-
- Stanislaus D, Pinter JH, Janovick JA, Conn PM. Mechanisms mediating multiple physiological responses to gonadotropin-releasing hormone. Mol Cell Endocrinol. 1998;144:1–10. - PubMed
-
- Brown P, McNeilly AS. Transcriptional regulation of pituitary gonadotrophin subunit genes. Rev Reprod. 1999;4:117–124. - PubMed
-
- Shacham S, Harris D, Ben-Shlomo H, Cohen I, Bonfil D, Przedecki F, Lewy H, Ashkenazi IE, Seger R, Naor Z. Mechanism of GnRH receptor signaling on gonadotropin release and gene expression in pituitary gonadotrophs. Vitam Horm. 2001;63:63–90. - PubMed
-
- Charlton HM, Halpin DM, Iddon C, Rosie R, Levy G, McDowell I, Megson A, Morris J, Bramwell A, Speight A, Ward B, Broadhead J, Davey-Smith G, Fink G. The effects of daily administration of single and multiple injections of gonadotropin releasing hormone on pituitary and gonadal function in the hypogonadal (HPG) mouse. Endocrinology. 1983;113:535–544. - PubMed
-
- Brown P, McNeilly J, Evans J, Crawford G, Walker M, Christian H, McNeilly A. Manipulating the in vivo mRNA expression profile of FSHβ to resemble that of LHβ does not promote a concomitant increase in intracellular storage of follicle-stimulating hormone. J Neuroendocrinol. 2001;13:50–62. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

