Genomic biodiversity, phylogenetics and coevolution in proteins
- PMID: 15130847
- PMCID: PMC2943949
Genomic biodiversity, phylogenetics and coevolution in proteins
Abstract
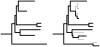
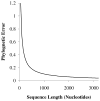
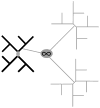
Comprehensive sampling of genomic biodiversity is fast becoming a reality for some genomic regions and complete organelle genomes. Genomic biodiversity is defined as large genomic sequences from many species, and here some recent work is reviewed that demonstrates the potential benefits of genomic biodiversity for molecular evolutionary analysis and phylogenetic reconstruction. This work shows that using likelihood-based approaches, taxon addition can dramatically improve phylogenetic reconstruction. Features or dynamics of the evolutionary process are much more easily inferred with large numbers of taxa, and large numbers are essential for discriminating differences in evolutionary patterns between sites. Accurate prediction of site-specific patterns can improve phylogenetic reconstruction by an amount equivalent to quadrupling sequence length. Genomic biodiversity is particularly central to research relating patterns of evolution, adaptation and coevolution to structural and functional features of proteins. Research on detecting coevolution between amino acid residues in proteins demonstrates a clear need for much greater numbers of closely related taxa to better discriminate site-specific patterns of interaction, and to allow more detailed analysis of coevolutionary interactions between subunits in protein complexes. It is argued that parsing out coevolutionary and other context-dependent substitution probabilities is essential for discriminating between coevolution and adaptation, and for more realistically modelling the evolution of proteins. Also reviewed is research that argues for increasing the efficiency of acquiring genomic biodiversity, and suggests that this might be done by simultaneously shotgun cloning and sequencing genomic mixtures from many species. Increased efficiency is a prerequisite if genomic biodiversity levels are to rapidly increase by orders of magnitude, and thus lead to dramatically improved understanding of interactions between protein structure, function and sequence evolution.
Figures
References
-
- Altschuh D, Lesk AM, Bloomer AC, Klug A. Correlation of coordinated amino acid substitutions with function in viruses related to tobacco mosaic virus. J Mol Biol. 1987;193:693–708. - PubMed
-
- Arctander P. Mitochondrial recombination? Science. 1999;284:2090–1. - PubMed
-
- Atchley WR, Wollenberg KR, Fitch WM, Terhalle W, Dress AW. Correlations among amino acid sites in bHLH protein domains: an information theoretic analysis. Mol Biol Evol. 2000;17:164–78. - PubMed
-
- Awadalla P, Eyre-Walker A, Maynard Smith J. Questioning evidence for recombination in human mitochondrial DNA: response. Science. 2000;288:1931a. - PubMed
-
- Awadalla P, Eyre-Walker A, Smith JM. Linkage disequilibrium and recombination in hominid mitochondrial DNA. Science. 1999;286:2524–5. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
