Viral RNA silencing suppressors inhibit the microRNA pathway at an intermediate step
- PMID: 15131083
- PMCID: PMC415642
- DOI: 10.1101/gad.1201204
Viral RNA silencing suppressors inhibit the microRNA pathway at an intermediate step
Erratum in
- Genes Dev. 2004 Jun 15;18(12):1510
Abstract
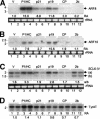
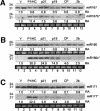
RNA silencing suppressors from different plant viruses are structurally diverse. In addition to inhibiting the antiviral silencing response to condition susceptibility, many suppressors are pathogenicity factors that cause disease or developmental abnormalities. Here, unrelated suppressors from multiple viruses were shown to inhibit microRNA (miRNA) activities and trigger an overlapping series of severe developmental defects in transgenic Arabidopsis thaliana. This suggests that interference with miRNA-directed processes may be a general feature contributing to pathogenicity of many viruses. A normally labile intermediate in the miRNA biogenesis/RNA-induced silencing complex (RISC) assembly pathway, miRNA*, accumulated specifically in the presence of suppressors (P1/HC-Pro, p21, or p19) that inhibited miRNA-guided cleavage of target mRNAs. Both p21 and p19, but not P1/HC-Pro, interacted with miRNA/miRNA* complexes and hairpin RNA-derived short interfering RNAs (siRNAs) in vivo. In addition, p21 bound to synthetic miRNA/miRNA* and siRNA duplexes in vitro. We propose that several different suppressors act by distinct mechanisms to inhibit the incorporation of small RNAs into active RISCs.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
