Performance assessment of DNA fragment sizing by high-sensitivity flow cytometry and pulsed-field gel electrophoresis
- PMID: 15131156
- PMCID: PMC404634
- DOI: 10.1128/JCM.42.5.1965-1976.2004
Performance assessment of DNA fragment sizing by high-sensitivity flow cytometry and pulsed-field gel electrophoresis
Abstract
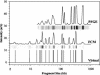
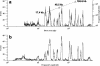
The sizing of restriction fragments is the chief analytical technique utilized in the production of DNA fingerprints. Few techniques have been able to compete with pulsed-field gel electrophoresis (PFGE), which is capable of discriminating among bacteria at species and strain levels by resolving restriction fragments. However, an ultrasensitive flow cytometer (FCM) developed in our lab has also demonstrated the ability to discriminate bacteria at species and strain levels. The abilities of FCM warrant a quantitative parallel comparison with PFGE to assess and evaluate the accuracy and precision of DNA fragment sizing by both techniques. Replicate samples of Staphylococcus aureus Mu50 were analyzed along with two clinical S. aureus isolates. The absolute fragment sizing accuracy was determined for PFGE (5% +/- 2%) and FCM (4% +/- 4%), with sequence-predicted Mu50 SmaI fragment sizes used as a reference. Precision was determined by simple arithmetic methods (relative standard deviation for PFGE [RSD(PFGE) ] = 3% +/- 2% and RSD(FCM) = 1.2% +/- 0.8%) as well as by the use of dendrograms derived from Dice coefficient-unweighted pair group method with arithmetic averages (UPGMA) and Pearson-UPGMA analyses. All quantitative measures of PFGE and FCM precision were equivalent, within error. The precision of both methods was not limited by any single sample preparation or analysis step that was tracked in this study. Additionally, we determined that the curve-based clustering of fingerprint data provided a more informative and useful assessment than did traditional band-based methods.
Figures




References
-
- Ambrose, W. P., H. Cai, P. M. Goodwin, J. H. Jett, R. C. Habbersett, E. J. Larson, W. K. Grace, J. H. Werner, and R. A. Keller. 2003. Flow cytometric sizing of DNA fragments, p. 239-270. In J. R. Lackowicz (ed.), Topics in fluorescence: DNA technology, vol. 7. Kluwer Academic/Plenum Publishers, New York, N.Y.
-
- Bai, J., Y. H. Liu, D. M. Lubman, and D. Siemieniak. 1994. Matrix-assisted laser-desorption ionization mass-spectrometry of restriction enzyme-digested plasmid DNA using an active nafion substrate. Rapid Commun. Mass Spectrom. 8:687-691. - PubMed
-
- Balaban, N., Y. Gov, A. Bitler, and J. R. Boelaert. 2003. Prevention of Staphylococcus aureus biofilm on dialysis catheters and adherence to human cells. Kidney Int. 63:340-345. - PubMed
-
- Birren, B., and E. Lai. 1993. Pulsed field gel electrophoresis: a practical guide. Academic Press, Inc., San Diego, Calif.
-
- Busch, U., and H. Nitschko. 1999. Methods for the differentiation of microorganisms. J. Chromatogr. B 722:263-278. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

