Reverse transcriptase PCR diagnostic assay for the coronavirus associated with severe acute respiratory syndrome
- PMID: 15131160
- PMCID: PMC404607
- DOI: 10.1128/JCM.42.5.1994-1999.2004
Reverse transcriptase PCR diagnostic assay for the coronavirus associated with severe acute respiratory syndrome
Abstract
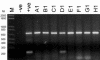
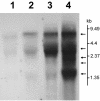
Recent outbreaks of severe acute respiratory syndrome (SARS) have spurred intense research efforts around the world to deal with the serious threat to health posed by this novel coronavirus. A rapid, reliable diagnostic assay is needed for monitoring the spread of the disease. Here we report a method for eliminating false-negative results and increasing test sensitivity, based on the hypothesis that the message encoded by the nucleocapsid (N) gene is the most abundant during viral infection. Nasopharyngeal aspirates and stool samples were obtained from suspected SARS patients with major clinical symptoms and a significant history of close contact with infected patients. Total RNAs were extracted in a 96-well format, together with pig kidney epithelial (PK-15) cells as an internal control for extraction efficiency. PCR inhibitors were removed by ethanol precipitation, and a PCR for the pig beta-actin gene was used as a positive control for all clinical samples. Samples were analyzed by a reverse transcriptase PCR assay. Northern blot analysis was performed to demonstrate differences in subgenomic transcripts of the virus, and a real-time quantitative PCR was employed to compare the sensitivities of two loci (1b and N). The detection rate of the assay reached 44.4% on day 9 after the onset of the disease. The diagnostic PCR amplifying the N gene gave an average of a 26.0% (6.3 to 60.0%) stronger intensity signal than that for the 1b gene. In conclusion, the nucleocapsid gene represents an additional sensitive molecular marker for the diagnosis of the SARS coronavirus and can be further adapted for use in a high-throughput platform assay.
Figures



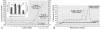
References
-
- Drosten, C., S. Günther, W. Presier, S. Werf, H. Brodt, S. Becker, et al. 2003. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 348:1967-1976. - PubMed
-
- Marra, M., S. Jones, C. Astell, R. Holt, A. Brooks-Wilson, et al. 2003. The complete genome sequence of SARS-associated coronavirus. Science 300:1377-1378. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

