Adenosine A1 and A2 receptors: structure--function relationships
- PMID: 1513184
- PMCID: PMC3448285
- DOI: 10.1002/med.2610120502
Adenosine A1 and A2 receptors: structure--function relationships
Figures
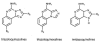

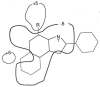
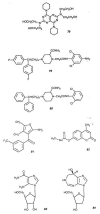


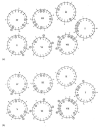


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

