Excision of Sleeping Beauty transposons: parameters and applications to gene therapy
- PMID: 15133768
- PMCID: PMC1865527
- DOI: 10.1002/jgm.486
Excision of Sleeping Beauty transposons: parameters and applications to gene therapy
Abstract
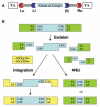
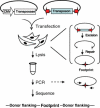
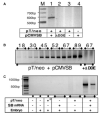
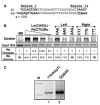
A major problem in gene therapy is the determination of the rates at which gene transfer has occurred. Our work has focused on applications of the Sleeping Beauty (SB) transposon system as a non-viral vector for gene therapy. Excision of a transposon from a donor molecule and its integration into a cellular chromosome are catalyzed by SB transposase. In this study, we used a plasmid-based excision assay to study the excision step of transposition. We used the excision assay to evaluate the importance of various sequences that border the sites of excision inside and outside the transposon in order to determine the most active sequences for transposition from a donor plasmid. These findings together with our previous results in transposase binding to the terminal repeats suggest that the sequences in the transposon-junction of SB are involved in steps subsequent to DNA binding but before excision, and that they may have a role in transposase-transposon interaction. We found that SB transposons leave characteristically different footprints at excision sites in different cell types, suggesting that alternative repair machineries operate in concert with transposition. Most importantly, we found that the rates of excision correlate with the rates of transposition. We used this finding to assess transposition in livers of mice that were injected with the SB transposon and transposase. The excision assay appears to be a relatively quick and easy method to optimize protocols for delivery of genes in SB transposons to mammalian chromosomes in living animals.
Copyright 2004 John Wiley & Sons, Ltd.
Figures


Similar articles
-
Sleeping Beauty, a wide host-range transposon vector for genetic transformation in vertebrates.J Mol Biol. 2000 Sep 8;302(1):93-102. doi: 10.1006/jmbi.2000.4047. J Mol Biol. 2000. PMID: 10964563
-
Sleeping beauty transposon-mediated gene therapy for prolonged expression.Adv Genet. 2005;54:189-232. doi: 10.1016/S0065-2660(05)54009-4. Adv Genet. 2005. PMID: 16096013 Review.
-
Counterselection and co-delivery of transposon and transposase functions for Sleeping Beauty-mediated transposition in cultured mammalian cells.Biosci Rep. 2004 Dec;24(6):577-94. doi: 10.1007/s10540-005-2793-9. Biosci Rep. 2004. PMID: 16158196
-
Gene transfer into genomes of human cells by the sleeping beauty transposon system.Mol Ther. 2003 Jul;8(1):108-17. doi: 10.1016/s1525-0016(03)00099-6. Mol Ther. 2003. PMID: 12842434
-
Sleeping Beauty Transposition.Microbiol Spectr. 2015 Apr;3(2):MDNA3-0042-2014. doi: 10.1128/microbiolspec.MDNA3-0042-2014. Microbiol Spectr. 2015. PMID: 26104705 Review.
Cited by
-
The impact of cHS4 insulators on DNA transposon vector mobilization and silencing in retinal pigment epithelium cells.PLoS One. 2012;7(10):e48421. doi: 10.1371/journal.pone.0048421. Epub 2012 Oct 26. PLoS One. 2012. PMID: 23110238 Free PMC article.
-
The Integration Preference of Sleeping Beauty at Non-TA Site Is Related to the Transposon End Sequences.Front Genet. 2021 Mar 10;12:639125. doi: 10.3389/fgene.2021.639125. eCollection 2021. Front Genet. 2021. PMID: 33777107 Free PMC article.
-
Sleeping Beauty Mouse Models of Cancer: Microenvironmental Influences on Cancer Genetics.Front Oncol. 2019 Jul 9;9:611. doi: 10.3389/fonc.2019.00611. eCollection 2019. Front Oncol. 2019. PMID: 31338332 Free PMC article. Review.
-
piggyBac-ing models and new therapeutic strategies.Trends Biotechnol. 2015 Sep;33(9):525-33. doi: 10.1016/j.tibtech.2015.06.009. Epub 2015 Jul 23. Trends Biotechnol. 2015. PMID: 26211958 Free PMC article. Review.
-
Evaluating the potential for undesired genomic effects of the piggyBac transposon system in human cells.Nucleic Acids Res. 2015 Feb 18;43(3):1770-82. doi: 10.1093/nar/gkv017. Epub 2015 Jan 20. Nucleic Acids Res. 2015. PMID: 25605795 Free PMC article.
References
-
- Plasterk RH. The Tc1/mariner transposon family. Curr Top Microbiol Immunol. 1996;204:125–143. - PubMed
-
- Plasterk RH, Izsvak Z, Ivics Z. Resident aliens: the Tc1/mariner superfamily of transposable elements. Trends Genet. 1999;15:326–332. - PubMed
-
- Ivics Z, Hackett PB, Plasterk RH, Izsvak Z. Molecular reconstruction of Sleeping Beauty, a Tc1-like transposon from fish, and its transposition in human cells. Cell. 1997;91:501–510. - PubMed
-
- Izsvak Z, Ivics Z, Plasterk RH. Sleeping Beauty, a wide host-range transposon vector for genetic transformation in vertebrates. J Mol Biol. 2000;302:93–102. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

