Identification of a novel protein 3a from severe acute respiratory syndrome coronavirus
- PMID: 15135062
- PMCID: PMC7126674
- DOI: 10.1016/j.febslet.2004.03.086
Identification of a novel protein 3a from severe acute respiratory syndrome coronavirus
Abstract
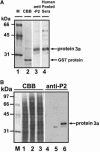
The open reading frame 3 of the severe acute respiratory syndrome coronavirus (SARS-CoV) genome encodes a predicted protein 3a, consisting of 274 amino acids, that lacks any significant similarities to any known protein. We generated specific antibodies against SARS protein 3a by using a synthetic peptide (P2) corresponding to amino acids 261-274 of the putative protein. Anti-P2 antibodies and the sera from SARS patients could specifically detect the recombinant SARS protein 3a expressed in Escherichia coli and in Vero E6 cells. Expression of SARS protein 3a was detected at 8-12 h after infection and reached a higher level after approximately 24 h in SARS-CoV-infected Vero E6 cells. Protein 3a was also detected in the alveolar lining pneumocytes and some intra-alveolar cells of a SARS-CoV-infected patient's lung specimen. Recombinant protein 3a expressed in Vero E6 cells and protein 3a in the SARS-CoV-infected cells was distributed over the cytoplasm in a fine punctate pattern with partly concentrated staining in the Golgi apparatus. Our study demonstrates that SARS-CoV indeed expresses a novel protein 3a, which is present only in SARS-CoV and not in other known CoVs.
Figures




References
-
- Ksiazek T.G., Erdman D., Goldsmith C.S., Zaki S.R., Peret T., Emery S., Tong S., Urbani C., Comer J.A., Lim W., N. Engl. J. Med, 348, (2003), 1953– 1966. - PubMed
-
- Holmes K.V., Enjuanes L., Science, 300, (2003), 1377– 1378. - PubMed
-
- Rota P.A., Oberste M.S., Monroe S.S., Nix W.A., Campagnoli R., Icenogle J.P., Penaranda S., Bankamp B., Maher K., Chen M.H., Science, 300, (2003), 1394– 1399. - PubMed
-
- Drosten C., Gunther S., Preiser W., van der Werf S., Brodt H.R., Becker S., Rabenau H., Panning M., Kolesnikova L., Fouchier R.A., N. Engl. J. Med, 348, (2003), 1967– 1976. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

