Functional interactions of RNA-capping enzyme with factors that positively and negatively regulate promoter escape by RNA polymerase II
- PMID: 15136722
- PMCID: PMC419647
- DOI: 10.1073/pnas.0401493101
Functional interactions of RNA-capping enzyme with factors that positively and negatively regulate promoter escape by RNA polymerase II
Abstract
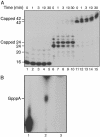
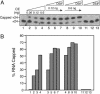
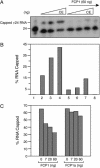
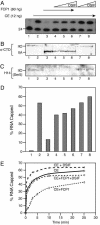
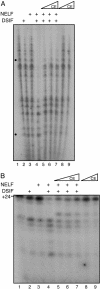
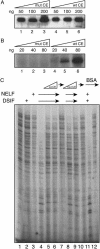
Capping of the 5' ends of nascent RNA polymerase II transcripts is the first pre-mRNA processing event in all eukaryotic cells. Capping enzyme (CE) is recruited to transcription complexes soon after initiation by the phosphorylation of Ser-5 of the carboxyl-terminal domain of the largest subunit of RNA polymerase II. Here, we analyze the role of CE in promoter clearance and its functional interactions with different factors that are involved in promoter clearance. FCP1-mediated dephosphorylation of the carboxyl-terminal domain results in a drastic decrease in cotranscriptional capping efficiency but is reversed by the presence of DRB sensitivity-inducing factor (DSIF). These results suggest involvement of DSIF in CE recruitment. Importantly, CE relieves transcriptional repression by the negative elongation factor, indicating a critical role of CE in the elongation checkpoint control mechanism during promoter clearance. This functional interaction between CE and the negative elongation factor documents a dynamic role of CE in promoter clearance beyond its catalytic activities.
Figures
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

