Nef binds p6* in GagPol during replication of human immunodeficiency virus type 1
- PMID: 15137387
- PMCID: PMC400368
- DOI: 10.1128/jvi.78.10.5311-5323.2004
Nef binds p6* in GagPol during replication of human immunodeficiency virus type 1
Abstract
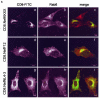
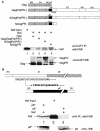
The atypical Nef protein (NefF12) from human immunodeficiency virus type 1 strain F12 (HIV-1(F12)) interferes with virion production and infectivity via a mysterious mechanism. The correlation of these effects with the unusual perinuclear subcellular localization of NefF12 suggested that the wild-type Nef protein could bind to assembly intermediates in late stages of viral replication. To test this hypothesis, Nef from HIV-1(NL4-3) was fused to an endoplasmic reticulum (ER) retention signal (NefKKXX). This mutant NefKKXX protein recapitulated fully the effects of NefF12 on on Gag processing and virion production, either alone or as a CD8 fusion protein. Importantly, the mutant NefKKXX protein also localized to the intermediate compartment, between the ER and the trans-Golgi network. Furthermore, Nef bound the GagPol polyprotein in vitro and in vivo. This binding mapped to the C-terminal flexible loop in Nef and the transframe p6* protein in GagPol. The significance of this interaction was demonstrated by a genetic assay in which the release of a mutant HIV-1 provirus lacking the PTAP motif in the late domain that no longer binds Tsg101 was rescued by a Nef.Tsg101 chimera. Importantly, this rescue as well as incorporation of Nef into HIV-1 virions correlated with the ability of Nef to interact with GagPol. Our data demonstrate that the retention of Nef in the intermediate compartment interferes with viral replication and suggest a new role for Nef in the production of HIV-1.
Figures



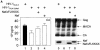
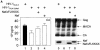
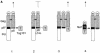
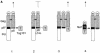
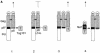
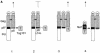
References
-
- Bresnahan, P. A., W. Yonemoto, S. Ferrell, D. Williams-Herman, R. Geleziunas, and W. C. Greene. 1998. A dileucine motif in HIV-1 Nef acts as an internalization signal for CD4 downregulation and binds the AP-1 clathrin adaptor. Curr. Biol. 8:1235-1238. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

