Mre11 assembles linear DNA fragments into DNA damage signaling complexes
- PMID: 15138496
- PMCID: PMC406388
- DOI: 10.1371/journal.pbio.0020110
Mre11 assembles linear DNA fragments into DNA damage signaling complexes
Erratum in
- PLoS Biol. 2004 Jun;2(6):876
Abstract
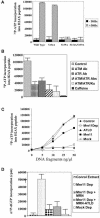
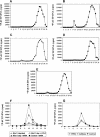
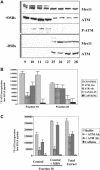
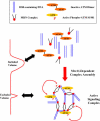
Mre11/Rad50/Nbs1 complex (MRN) is essential to suppress the generation of double-strand breaks (DSBs) during DNA replication. MRN also plays a role in the response to DSBs created by DNA damage. Hypomorphic mutations in Mre11 (which causes an ataxia-telangiectasia-like disease [ATLD]) and mutations in the ataxia-telangiectasia-mutated (ATM) gene lead to defects in handling damaged DNA and to similar clinical and cellular phenotypes. Using Xenopus egg extracts, we have designed a simple assay to define the biochemistry of Mre11. MRN is required for efficient activation of the DNA damage response induced by DSBs. We isolated a high molecular weight DNA damage signaling complex that includes MRN, damaged DNA molecules, and activated ATM. Complex formation is partially dependent upon Zn(2+) and requires an intact Mre11 C-terminal domain that is deleted in some ATLD patients. The ATLD truncation can still perform the role of Mre11 during replication. Our work demonstrates the role of Mre11 in assembling DNA damage signaling centers that are reminiscent of irradiation-induced foci. It also provides a molecular explanation for the similarities between ataxia-telangiectasia (A-T) and ATLD.
Conflict of interest statement
The authors have declared that no conflicts of interest exist.
Figures

References
-
- Abraham RT. Cell cycle checkpoint signaling through the ATM and ATR kinases. Genes Dev. 2001;15:2177–2196. - PubMed
-
- Andegeko Y, Moyal L, Mittelman L, Tsarfaty I, Shiloh Y, et al. Nuclear retention of ATM at sites of DNA double strand breaks. J Biol Chem. 2001;276:38224–38230. - PubMed
-
- Aten JA, Stap J, Krawczyk PM, van Oven CH, Hoebe RA, et al. Dynamics of DNA double-strand breaks revealed by clustering of damaged chromosome domains. Science. 2004;303:92–95. - PubMed
-
- Bakkenist CJ, Kastan MB. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature. 2003;421:499–506. - PubMed
-
- Burma S, Chen BP, Murphy M, Kurimasa A, Chen DJ. ATM phosphorylates histone H2AX in response to DNA double-strand breaks. J Biol Chem. 2001;276:42462–42467. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

