Ras and Gpa2 mediate one branch of a redundant glucose signaling pathway in yeast
- PMID: 15138498
- PMCID: PMC406390
- DOI: 10.1371/journal.pbio.0020128
Ras and Gpa2 mediate one branch of a redundant glucose signaling pathway in yeast
Abstract
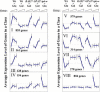
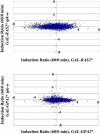
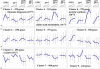
Addition of glucose to starved yeast cells elicits a dramatic restructuring of the transcriptional and metabolic state of the cell. While many components of the signaling network responsible for this response have been identified, a comprehensive view of this network is lacking. We have used global analysis of gene expression to assess the roles of the small GTP-binding proteins, Ras2 and Gpa2, in mediating the transcriptional response to glucose. We find that 90% of the transcriptional changes in the cell attendant on glucose addition are recapitulated by activation of Ras2 or Gpa2. In addition, we find that protein kinase A (PKA) mediates all of the Ras2 and Gpa2 transcriptional effects. However, we also find that most of the transcriptional effects of glucose addition to wild-type cells are retained in strains containing a PKA unresponsive to changes in cAMP levels. Thus, most glucose-responsive genes are regulated redundantly by a Ras/PKA-dependent pathway and by one or more PKA-independent pathways. Computational analysis extracted RRPE/PAC as the major response element for Ras and glucose regulation and revealed additional response elements mediating glucose and Ras regulation. These studies provide a paradigm for extracting the topology of signal transduction pathways from expression data.
Conflict of interest statement
The authors have declared that no conflicts of interest exist.
Figures



References
-
- Broach JR, Deschenes RJ. The function of ras genes in Saccharomyces cerevisiae . Adv Cancer Res. 1990;54:79–139. - PubMed
-
- Cameron S, Levin L, Zoller M, Wigler M. cAMP-independent control of sporulation, glycogen metabolism, and heat shock resistance in S. cerevisiae . Cell. 1988;53:555–566. - PubMed
-
- Carlson M. Glucose repression in yeast. Curr Opin Microbiol. 1999;2:202–207. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

