Systems level insights into the stress response to UV radiation in the halophilic archaeon Halobacterium NRC-1
- PMID: 15140832
- PMCID: PMC419780
- DOI: 10.1101/gr.1993504
Systems level insights into the stress response to UV radiation in the halophilic archaeon Halobacterium NRC-1
Abstract
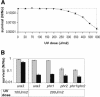
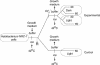
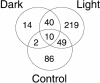
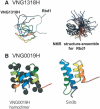
We report a remarkably high UV-radiation resistance in the extremely halophilic archaeon Halobacterium NRC-1 withstanding up to 110 J/m2 with no loss of viability. Gene knockout analysis in two putative photolyase-like genes (phr1 and phr2) implicated only phr2 in photoreactivation. The UV-response was further characterized by analyzing simultaneously, along with gene function and protein interactions inferred through comparative genomics approaches, mRNA changes for all 2400 genes during light and dark repair. In addition to photoreactivation, three other putative repair mechanisms were identified including d(CTAG) methylation-directed mismatch repair, four oxidative damage repair enzymes, and two proteases for eliminating damaged proteins. Moreover, a UV-induced down-regulation of many important metabolic functions was observed during light repair and seems to be a phenomenon shared by all three domains of life. The systems analysis has facilitated the assignment of putative functions to 26 of 33 key proteins in the UV response through sequence-based methods and/or similarities of their predicted three-dimensional structures to known structures in the PDB. Finally, the systems analysis has raised, through the integration of experimentally determined and computationally inferred data, many experimentally testable hypotheses that describe the metabolic and regulatory networks of Halobacterium NRC-1.
Copyright 2004 Cold Spring Harbor Laboratory Press
Figures


References
-
- Amillet, J.M., Buisson, N., and Labbe-Bois, R. 1995. Positive and negative elements involved in the differential regulation by heme and oxygen of the HEM13 gene (coproporphyrinogen oxidase) in Saccharomyces cerevisiae. Curr. Genet. 28: 503–511. - PubMed
-
- Bonneau, R., Tsai, J., Ruczinski, I., Chivian, D., Rohl, C., Strauss, C.E., and Baker, D. 2001. Rosetta in CASP4: Progress in ab initio protein structure prediction. Proteins Suppl: 119–126. - PubMed
WEB SITE REFERENCES
-
- http://bioinfo.pl/meta/; A Meta server provides access to various fold recognition and local structure prediction methods. This server also provides translation of the standard formats like PDB, CASP, or PIR, and is coupled to several consensus servers such as pcons.
-
- http://www.sbc.su.se/∼arne/pcons/; A consensus fold recognition predictor.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
