T-cell epitopes in severe acute respiratory syndrome (SARS) coronavirus spike protein elicit a specific T-cell immune response in patients who recover from SARS
- PMID: 15140958
- PMCID: PMC415819
- DOI: 10.1128/JVI.78.11.5612-5618.2004
T-cell epitopes in severe acute respiratory syndrome (SARS) coronavirus spike protein elicit a specific T-cell immune response in patients who recover from SARS
Erratum in
- J Virol. 2004 Jul;78(14):7861. Yang Huang-Hua [corrected to Yang Huang-Hao]
Abstract
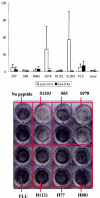
The immunogenicity of HLA-A2-restricted T-cell epitopes in the S protein of the Severe acute respiratory syndrome coronavirus (SARS-CoV) and of human coronavirus strain 229e (HCoV-229e) was analyzed for the elicitation of a T-cell immune response in donors who had fully recovered from SARS-CoV infection. We employed online database analysis to compare the differences in the amino acid sequences of the homologous T epitopes of HCoV-229e and SARS-CoV. The identified T-cell epitope peptides were synthesized, and their binding affinities for HLA-A2 were validated and compared in the T2 cell system. The immunogenicity of all these peptides was assessed by using T cells obtained from donors who had fully recovered from SARS-CoV infection and from healthy donors with no history of SARS-CoV infection. HLA-A2 typing by indirect immunofluorescent antibody staining showed that 51.6% of SARS-CoV-infected patients were HLA-A2 positive. Online database analysis and the T2 cell binding test disclosed that the number of HLA-A2-restricted immunogenic epitopes of the S protein of SARS-CoV was decreased or even lost in comparison with the homologous sequences of the S protein of HCoV-229e. Among the peptides used in the study, the affinity of peptides from HCoV-229e (H77 and H881) and peptides from SARS-CoV (S978 and S1203) for binding to HLA-A2 was higher than that of other sequences. The gamma interferon (IFN-gamma) release Elispot assay revealed that only SARS-CoV-specific peptides S1203 and S978 induced a high frequency of IFN-gamma-secreting T-cell response in HLA-A2(+) donors who had fully recovered from SARS-CoV infection; such a T-cell epitope-specific response was not observed in HLA-A2(+) healthy donors or in HLA-A2(-) donors who had been infected with SARS-CoV after full recovery. Thus, T-cell epitopes S1203 and S978 are immunogenic and elicit an overt specific T-cell response in HLA-A2(+) SARS-CoV-infected patients.
Figures

References
-
- Bartulewic, D., A. Markowska, S. Wolczynski, M. Dabrowska, and A. Rozanski. 2000. Molecular modelling, synthesis and antitumour activity of carbocyclic analogues of netropsin and distamycin-new carriers of alkylating elements. Acta Biochim. Pol. 47:23-35. - PubMed
-
- Cerundolo, V., J. Alexander, K. Anderson, C. Lamb, P. Cresswell, A. McMichael, F. Gotch, and A. Townsend. 1990. Presentation of viral antigen controlled by a gene in the major histocompatibility complex. Nature (London) 345:449-452. - PubMed
-
- Gnjatic, S., D. Atanackovic, M. Matsuo, E. Jager, S. Y. Lee, D. Valmori, Y. T. Chen, G., Ritter, A. Knuth, and L. J. Old. 2003. Cross-presentation of HLA class I epitopes from exogenous N. Y.-ESO-1 polypeptides by nonprofessional APCs. J. Immunol. 170:1191-1196. - PubMed
-
- Gricks, C. S., E. Rawlings, L. Foroni, J. A. Madrigal, and P. L. Amlot. 2001. Somatically mutated regions of immunoglobulin on human b-cell lymphomas code for peptides that bind to autologous major histocompatibility complex class I, providing a potential target for cytotoxic T cells. Cancer Res. 61:5145-5152. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

