Processing of a pestivirus protein by a cellular protease specific for light chain 3 of microtubule-associated proteins
- PMID: 15140988
- PMCID: PMC415803
- DOI: 10.1128/JVI.78.11.5900-5912.2004
Processing of a pestivirus protein by a cellular protease specific for light chain 3 of microtubule-associated proteins
Abstract
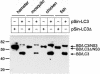
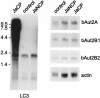
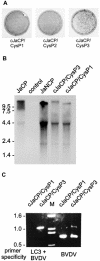
The genome of the cytopathogenic (cp) bovine viral diarrhea virus (BVDV) JaCP contains a cellular insertion coding for light chain 3 (LC3) of microtubule-associated proteins, the mammalian homologue of yeast Aut7p/Apg8p. The cellular insertion induces cp BVDV-specific processing of the viral polyprotein by a cellular cysteine protease homologous to the known yeast protease Aut2p/Apg4p. Three candidate bovine protease genes were identified on the basis of the sequence similarity of their products with the Saccharomyces cerevisiae enzyme. The search for a system for functional testing of these putative LC3-specific proteases revealed that the components involved in this processing have been highly conserved during evolution, so that the substrate derived from a mammalian virus is processed in cells of mammalian, avian, fish, and insect origin, as well as in rabbit reticulocyte lysate, but not in wheat germ extracts. Moreover, two of these proteases and a homologous protein from chickens were able to rescue the defect of a yeast AUT2 deletion mutant. In coexpression experiments with yeast and wheat germ extracts one of the bovine proteases and the corresponding enzyme from chickens were able to process the viral polyprotein containing LC3. Northern blots showed that bovine viral diarrhea virus infection of cells has no significant influence on the expression of either LC3 or its protease, bAut2B2. However, LC3-specific processing of the viral polyprotein containing the cellular insertion is essential for replication of the virus since mutants with changes in the LC3 insertion significantly affecting processing at the LC3/NS3 site were not viable.
Figures


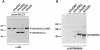




Similar articles
-
Insertion of a sequence encoding light chain 3 of microtubule-associated proteins 1A and 1B in a pestivirus genome: connection with virus cytopathogenicity and induction of lethal disease in cattle.J Virol. 1998 May;72(5):4139-48. doi: 10.1128/JVI.72.5.4139-4148.1998. J Virol. 1998. PMID: 9557703 Free PMC article.
-
Temporal modulation of an autoprotease is crucial for replication and pathogenicity of an RNA virus.J Virol. 2004 Oct;78(19):10765-75. doi: 10.1128/JVI.78.19.10765-10775.2004. J Virol. 2004. PMID: 15367643 Free PMC article.
-
Cellular sequences in pestivirus genomes encoding gamma-aminobutyric acid (A) receptor-associated protein and Golgi-associated ATPase enhancer of 16 kilodaltons.J Virol. 2002 Dec;76(24):13069-76. doi: 10.1128/jvi.76.24.13069-13076.2002. J Virol. 2002. PMID: 12438634 Free PMC article.
-
Non-structural proteins of bovine viral diarrhea virus.Virus Genes. 2022 Dec;58(6):491-500. doi: 10.1007/s11262-022-01914-8. Epub 2022 May 25. Virus Genes. 2022. PMID: 35614328 Free PMC article. Review.
-
Bovine viral diarrhea virus proteins and their antigenic analyses.Arch Virol Suppl. 1991;3:29-40. doi: 10.1007/978-3-7091-9153-8_4. Arch Virol Suppl. 1991. PMID: 9210923 Review.
Cited by
-
Autophagy and viruses: adversaries or allies?J Innate Immun. 2013;5(5):480-93. doi: 10.1159/000346388. Epub 2013 Jan 31. J Innate Immun. 2013. PMID: 23391695 Free PMC article. Review.
-
Coxsackievirus can exploit LC3 in both autophagy-dependent and -independent manners in vivo.Autophagy. 2015;11(8):1389-407. doi: 10.1080/15548627.2015.1063769. Autophagy. 2015. PMID: 26090585 Free PMC article.
-
Cross-talking between autophagy and viral infection in mammalian cells.Front Biol (Beijing). 2010;5(6):507-515. doi: 10.1007/s11515-010-0760-8. Epub 2010 Dec 11. Front Biol (Beijing). 2010. PMID: 32215004 Free PMC article. Review.
-
Autophagy in viral replication and pathogenesis.Mol Cells. 2010 Jan;29(1):1-7. doi: 10.1007/s10059-010-0014-2. Epub 2010 Jan 8. Mol Cells. 2010. PMID: 20077024 Free PMC article. Review.
-
Involvement of autophagy in viral infections: antiviral function and subversion by viruses.J Mol Med (Berl). 2007 Aug;85(8):811-23. doi: 10.1007/s00109-007-0173-6. Epub 2007 Mar 6. J Mol Med (Berl). 2007. PMID: 17340132 Free PMC article. Review.
References
-
- Ausubel, F. M., R. Brent, R. E. Kingston, and D. D. Moore (ed.). 1987. Current protocols in molecular biology. Greene Publishing Associates, New York, N.Y.
-
- Baroth, M., M. Orlich, H. J. Thiel, and P. Becher. 2000. Insertion of cellular NEDD8 coding sequences in a pestivirus. Virology 278:456-466. - PubMed
-
- Barth, H., and M. Thumm. 2001. A genomic screen identifies AUT8 as a novel gene essential for autophagy in the yeast Saccharomyces cerevisiae. Gene 274:151-156. - PubMed
-
- Becher, P., A. D. Shannon, N. Tautz, and H.-J. Thiel. 1994. Molecular characterization of border disease virus, a pestivirus from sheep. Virology 198:542-551. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

