Recombinant Sendai virus expressing the G glycoprotein of respiratory syncytial virus (RSV) elicits immune protection against RSV
- PMID: 15141002
- PMCID: PMC415788
- DOI: 10.1128/JVI.78.11.6043-6047.2004
Recombinant Sendai virus expressing the G glycoprotein of respiratory syncytial virus (RSV) elicits immune protection against RSV
Abstract
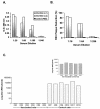
Although RSV causes serious pediatric respiratory disease, an effective vaccine does not exist. To capture the strengths of a live virus vaccine, we have used the murine parainfluenza virus type 1 (Sendai virus [SV]) as a xenogeneic vector to deliver the G glycoprotein of RSV. It was previously shown (J. L. Hurwitz, K. F. Soike, M. Y. Sangster, A. Portner, R. E. Sealy, D. H. Dawson, and C. Coleclough, Vaccine 15:533-540, 1997) that intranasal SV protected African green monkeys from challenge with the related human parainfluenza virus type 1 (hPIV1), and SV has advanced to clinical trials as a vaccine for hPIV1 (K. S. Slobod, J. L. Shenep, J. Lujan-Zilbermann, K. Allison, B. Brown, R. A. Scroggs, A. Portner, C. Coleclough, and J. L. Hurwitz, Vaccine, in press). Recombinant SV expressing RSV G glycoprotein was prepared by using reverse genetics, and intranasal inoculation of cotton rats elicited RSV-specific antibody and elicited protection from RSV challenge. RSV G-recombinant SV is thus a promising live virus vaccine candidate for RSV.
Figures


References
-
- Anderson, L. J., R. A. Parker, and R. L. Strikas. 1990. Association between respiratory syncytial virus outbreaks and lower respiratory tract deaths of infants and young children. J. Infect. Dis. 161:640-646. - PubMed
-
- Connors, M., A. B. Kulkarni, C.-Y. Firestone, K. L. Holmes, H. C. Morse, A. V. Sotnikov, and B. R. Murphy. 1992. Pulmonary histopathology induced by respiratory syncytial virus (RSV) challenge of formalin-inactivated RSV-immunized BALB/c mice is abrogated by depletion of CD4+ T cells. J. Virol. 66:7444-7451. - PMC - PubMed
-
- Crowe, J. E., Jr., P. L. Collins, W. T. London, R. M. Chanock, and B. R. Murphy. 1993. A comparison in chimpanzees of the immunogenicity and efficacy of live attenuated respiratory syncytial virus (RSV) temperature-sensitive mutant vaccines and vaccinia virus recombinants that express the surface glycoproteins of RSV. Vaccine 11:1395-1404. - PubMed
-
- Fulginiti, V. A., J. J. Eller, O. F. Sieber, J. W. Joyner, M. Minamitani, and G. Meiklejohn. 1969. I. A field trial of two inactivated respiratory virus vaccines; an aqueous trivalent parainfluenza virus vaccine and an alum-precipitated respiratory syncytial virus vaccine. Am. J. Epidemiol. 89:435-448. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

