Reverse genetic analysis of the transcription regulatory sequence of the coronavirus transmissible gastroenteritis virus
- PMID: 15141005
- PMCID: PMC415797
- DOI: 10.1128/JVI.78.11.6061-6066.2004
Reverse genetic analysis of the transcription regulatory sequence of the coronavirus transmissible gastroenteritis virus
Abstract
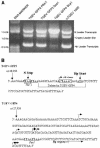
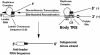
Coronavirus discontinuous transcription uses a highly conserved sequence (CS) in the joining of leader and body RNAs. Using a full-length infectious construct of transmissable gastroenteritis virus, the present study demonstrates that subgenomic transcription is heavily influenced by upstream flanking sequences and supports a mechanism of transcription attenuation that is regulated in part by a larger domain composed of primarily upstream flanking sequences which select appropriately positioned CS elements for synthesis of subgenomic RNAs.
Figures

Similar articles
-
Transcription regulatory sequences and mRNA expression levels in the coronavirus transmissible gastroenteritis virus.J Virol. 2002 Feb;76(3):1293-308. doi: 10.1128/jvi.76.3.1293-1308.2002. J Virol. 2002. PMID: 11773405 Free PMC article.
-
Role of nucleotides immediately flanking the transcription-regulating sequence core in coronavirus subgenomic mRNA synthesis.J Virol. 2005 Feb;79(4):2506-16. doi: 10.1128/JVI.79.4.2506-2516.2005. J Virol. 2005. PMID: 15681451 Free PMC article.
-
Sequence motifs involved in the regulation of discontinuous coronavirus subgenomic RNA synthesis.J Virol. 2004 Jan;78(2):980-94. doi: 10.1128/jvi.78.2.980-994.2004. J Virol. 2004. PMID: 14694129 Free PMC article.
-
Expression of transcriptional units using transmissible gastroenteritis coronavirus derived minigenomes and full-length cDNA clones.Adv Exp Med Biol. 2001;494:447-51. doi: 10.1007/978-1-4615-1325-4_65. Adv Exp Med Biol. 2001. PMID: 11774506 Review. No abstract available.
-
Coronavirus reverse genetics and development of vectors for gene expression.Curr Top Microbiol Immunol. 2005;287:161-97. doi: 10.1007/3-540-26765-4_6. Curr Top Microbiol Immunol. 2005. PMID: 15609512 Free PMC article. Review.
Cited by
-
Functional and genetic analysis of coronavirus replicase-transcriptase proteins.PLoS Pathog. 2005 Dec;1(4):e39. doi: 10.1371/journal.ppat.0010039. Epub 2005 Dec 9. PLoS Pathog. 2005. PMID: 16341254 Free PMC article. Review.
-
Coronaviruses: an RNA proofreading machine regulates replication fidelity and diversity.RNA Biol. 2011 Mar-Apr;8(2):270-9. doi: 10.4161/rna.8.2.15013. Epub 2011 Mar 1. RNA Biol. 2011. PMID: 21593585 Free PMC article. Review.
-
The molecular biology of coronaviruses.Adv Virus Res. 2006;66:193-292. doi: 10.1016/S0065-3527(06)66005-3. Adv Virus Res. 2006. PMID: 16877062 Free PMC article. Review.
-
Torovirus non-discontinuous transcription: mutational analysis of a subgenomic mRNA promoter.J Virol. 2005 Jul;79(13):8275-81. doi: 10.1128/JVI.79.13.8275-8281.2005. J Virol. 2005. PMID: 15956573 Free PMC article.
-
Reverse genetic manipulation of the overlapping coding regions for structural proteins of the type II porcine reproductive and respiratory syndrome virus.Virology. 2009 Jan 5;383(1):22-31. doi: 10.1016/j.virol.2008.09.013. Epub 2008 Nov 5. Virology. 2009. PMID: 18977502 Free PMC article.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

