Mismatch cleavage by single-strand specific nucleases
- PMID: 15141034
- PMCID: PMC419476
- DOI: 10.1093/nar/gkh599
Mismatch cleavage by single-strand specific nucleases
Abstract
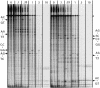
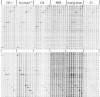
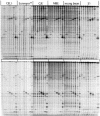
We have investigated the ability of single-strand specific (sss) nucleases from different sources to cleave single base pair mismatches in heteroduplex DNA templates used for mutation and single-nucleotide polymorphism analysis. The TILLING (Targeting Induced Local Lesions IN Genomes) mismatch cleavage protocol was used with the LI-COR gel detection system to assay cleavage of amplified heteroduplexes derived from a variety of induced mutations and naturally occurring polymorphisms. We found that purified nucleases derived from celery (CEL I), mung bean sprouts and Aspergillus (S1) were able to specifically cleave nearly all single base pair mismatches tested. Optimal nicking of heteroduplexes for mismatch detection was achieved using higher pH, temperature and divalent cation conditions than are routinely used for digestion of single-stranded DNA. Surprisingly, crude plant extracts performed as well as the highly purified preparations for this application. These observations suggest that diverse members of the S1 family of sss nucleases act similarly in cleaving non-specifically at bulges in heteroduplexes, and single-base mismatches are the least accessible because they present the smallest single-stranded region for enzyme binding. We conclude that a variety of sss nucleases and extracts can be effectively used for high-throughput mutation and polymorphism discovery.
Figures

References
-
- Desai N.A. and Shankar,V. (2003) Single-strand-specific nucleases. FEMS Microbiol. Rev., 26, 457–491. - PubMed
-
- Wiegand R.C., Godson,N. and Radding,C.M. (1975) Specificity of the S1 nuclease from Aspergillus oryzae. J. Biol. Chem., 250, 8848–8855. - PubMed
-
- Berk A.J. (1989) Characterization of RNA molecules by S1 nuclease analysis. Methods Enzymol., 180, 334–347. - PubMed

