Structural basis for the selective inhibition of JNK1 by the scaffolding protein JIP1 and SP600125
- PMID: 15141161
- PMCID: PMC419904
- DOI: 10.1038/sj.emboj.7600212
Structural basis for the selective inhibition of JNK1 by the scaffolding protein JIP1 and SP600125
Abstract
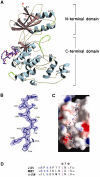
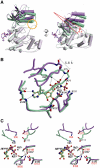
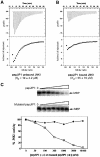
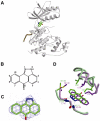
The c-jun N-terminal kinase (JNK) signaling pathway is regulated by JNK-interacting protein-1 (JIP1), which is a scaffolding protein assembling the components of the JNK cascade. Overexpression of JIP1 deactivates the JNK pathway selectively by cytoplasmic retention of JNK and thereby inhibits gene expression mediated by JNK, which occurs in the nucleus. Here, we report the crystal structure of human JNK1 complexed with pepJIP1, the peptide fragment of JIP1, revealing its selectivity for JNK1 over other MAPKs and the allosteric inhibition mechanism. The van der Waals contacts by the three residues (Pro157, Leu160, and Leu162) of pepJIP1 and the hydrogen bonding between Glu329 of JNK1 and Arg156 of pepJIP1 are critical for the selective binding. Binding of the peptide also induces a hinge motion between the N- and C-terminal domains of JNK1 and distorts the ATP-binding cleft, reducing the affinity of the kinase for ATP. In addition, we also determined the ternary complex structure of pepJIP1-bound JNK1 complexed with SP600125, an ATP-competitive inhibitor of JNK, providing the basis for the JNK specificity of the compound.
Figures



Similar articles
-
Understanding the specificity of a docking interaction between JNK1 and the scaffolding protein JIP1.J Phys Chem B. 2011 Feb 17;115(6):1491-502. doi: 10.1021/jp1073522. Epub 2011 Jan 25. J Phys Chem B. 2011. PMID: 21261310 Free PMC article.
-
Bipartite binding of the intrinsically disordered scaffold protein JIP1 to the kinase JNK1.Proc Natl Acad Sci U S A. 2025 Mar 4;122(9):e2419915122. doi: 10.1073/pnas.2419915122. Epub 2025 Feb 25. Proc Natl Acad Sci U S A. 2025. PMID: 39999166 Free PMC article.
-
Licochalcone A, a natural inhibitor of c-Jun N-terminal kinase 1.Cancer Prev Res (Phila). 2014 Jan;7(1):139-49. doi: 10.1158/1940-6207.CAPR-13-0117. Epub 2013 Nov 19. Cancer Prev Res (Phila). 2014. PMID: 24253317
-
Identification of small-molecule inhibitors of the JIP-JNK interaction.Biochem J. 2009 May 13;420(2):283-94. doi: 10.1042/BJ20081899. Biochem J. 2009. PMID: 19243309
-
Inhibitors of c-Jun N-terminal kinases: JuNK no more?Biochim Biophys Acta. 2008 Jan;1784(1):76-93. doi: 10.1016/j.bbapap.2007.09.013. Epub 2007 Oct 11. Biochim Biophys Acta. 2008. PMID: 17964301 Free PMC article. Review.
Cited by
-
Interface analysis of the complex between ERK2 and PTP-SL.PLoS One. 2009;4(5):e5432. doi: 10.1371/journal.pone.0005432. Epub 2009 May 8. PLoS One. 2009. PMID: 19424502 Free PMC article.
-
Targeting innate immunity protein kinase signalling in inflammation.Nat Rev Drug Discov. 2009 Jun;8(6):480-99. doi: 10.1038/nrd2829. Nat Rev Drug Discov. 2009. PMID: 19483709 Review.
-
Suppression of cytokine signaling by SOCS3: characterization of the mode of inhibition and the basis of its specificity.Immunity. 2012 Feb 24;36(2):239-50. doi: 10.1016/j.immuni.2011.12.015. Immunity. 2012. PMID: 22342841 Free PMC article.
-
Selective killing of p53-deficient cancer cells by SP600125.EMBO Mol Med. 2012 Jun;4(6):500-14. doi: 10.1002/emmm.201200228. Epub 2012 Mar 21. EMBO Mol Med. 2012. PMID: 22438244 Free PMC article.
-
Functional divergence caused by mutations in an energetic hotspot in ERK2.Proc Natl Acad Sci U S A. 2019 Jul 30;116(31):15514-15523. doi: 10.1073/pnas.1905015116. Epub 2019 Jul 11. Proc Natl Acad Sci U S A. 2019. PMID: 31296562 Free PMC article.
References
-
- Barr RK, Kendrick TS, Bogoyevitch MA (2002) Identification of the critical features of a small peptide inhibitor of JNK activity. J Biol Chem 277: 10987–10997 - PubMed
-
- Bonny C, Oberson A, Negri S, Sauser C, Schorderet DF (2001) Cell-permeable peptide inhibitors of JNK: novel blockers of beta-cell death. Diabetes 50: 77–82 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

