Organ distribution of severe acute respiratory syndrome (SARS) associated coronavirus (SARS-CoV) in SARS patients: implications for pathogenesis and virus transmission pathways
- PMID: 15141376
- PMCID: PMC7167761
- DOI: 10.1002/path.1560
Organ distribution of severe acute respiratory syndrome (SARS) associated coronavirus (SARS-CoV) in SARS patients: implications for pathogenesis and virus transmission pathways
Abstract
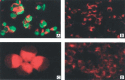
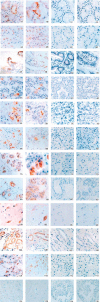
We previously identified the major pathological changes in the respiratory and immune systems of patients who died of severe acute respiratory syndrome (SARS) but gained little information on the organ distribution of SARS-associated coronavirus (SARS-CoV). In the present study, we used a murine monoclonal antibody specific for SARS-CoV nucleoprotein, and probes specific for a SARS-CoV RNA polymerase gene fragment, for immunohistochemistry and in situ hybridization, respectively, to detect SARS-CoV systematically in tissues from patients who died of SARS. SARS-CoV was found in lung, trachea/bronchus, stomach, small intestine, distal convoluted renal tubule, sweat gland, parathyroid, pituitary, pancreas, adrenal gland, liver and cerebrum, but was not detected in oesophagus, spleen, lymph node, bone marrow, heart, aorta, cerebellum, thyroid, testis, ovary, uterus or muscle. These results suggest that, in addition to the respiratory system, the gastrointestinal tract and other organs with detectable SARS-CoV may also be targets of SARS-CoV infection. The pathological changes in these organs may be caused directly by the cytopathic effect mediated by local replication of the SARS-CoV; or indirectly as a result of systemic responses to respiratory failure or the harmful immune response induced by viral infection. In addition to viral spread through a respiratory route, SARS-CoV in the intestinal tract, kidney and sweat glands may be excreted via faeces, urine and sweat, thereby leading to virus transmission. This study provides important information for understanding the pathogenesis of SARS-CoV infection and sheds light on possible virus transmission pathways. This data will be useful for designing new strategies for prevention and treatment of SARS.
Copyright 2004 Pathological Society of Great Britain and Ireland. Published by John Wiley & Sons, Ltd.
Figures

References
-
- WHO . Summary of probable SARS cases with onset of illness from 1 November 2002 to 31 July 2003. http://www.who.int [2003].
-
- Ksiazek TG, Erdman D, Goldsmith CS, et al. A novel coronavirus associated with severe acute respiratory syndrome. N Engl J Med 2003; 348: 1953–1966. - PubMed
-
- Drosten C, Gunther S, Preiser W, et al. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N Engl J Med 2003; 348: 1967–1976. - PubMed
-
- Marra MA, Jones SJM, Astell CR, et al. The genome sequence of the SARS‐associated coronavirus. Science 2003; 300: 1399–1404. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

