The Rpd3-Sin3 histone deacetylase regulates replication timing and enables intra-S origin control in Saccharomyces cerevisiae
- PMID: 15143171
- PMCID: PMC416400
- DOI: 10.1128/MCB.24.11.4769-4780.2004
The Rpd3-Sin3 histone deacetylase regulates replication timing and enables intra-S origin control in Saccharomyces cerevisiae
Abstract
The replication of eukaryotic genomes follows a temporally staged program, in which late origin firing often occurs within domains of altered chromatin structure(s) and silenced genes. Histone deacetylation functions in gene silencing in some late-replicating regions, prompting an investigation of the role of histone deacetylation in replication timing control in Saccharomyces cerevisiae. Deletion of the histone deacetylase Rpd3 or its interacting partner Sin3 caused early activation of late origins at internal chromosomal loci but did not alter the initiation timing of early origins or a late-firing, telomere-proximal origin. By delaying initiation relative to the earliest origins, Rpd3 enables regulation of late origins by the intra-S replication checkpoint. RPD3 deletion suppresses the slow S phase of clb5Delta cells by enabling late origins to fire earlier, suggesting that Rpd3 modulates the initiation timing of many origins throughout the genome. Examination of factors such as Ume6 that function together with Rpd3 in transcriptional repression indicates that Rpd3 regulates origin initiation timing independently of its role in transcriptional repression. This supports growing evidence that for much of the S. cerevisiae genome transcription and replication timing are not linked.
Figures


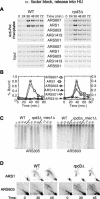

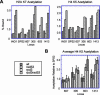

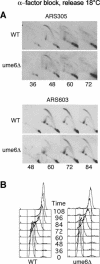
References
-
- Aparicio, O. M. 1999. Characterization of proteins bound to chromatin by immunoprecipitation from whole-cell extracts, p. 21.3.1-21.3.12. In F. M. Ausubel, R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.), Current protocols in molecular biology, vol. 4. John Wiley and Sons, Inc., New York, N.Y.
-
- Aparicio, O. M., D. M. Weinstein, and S. P. Bell. 1997. Components and dynamics of DNA replication complexes in S. cerevisiae: redistribution of MCM proteins and Cdc45p during S phase. Cell 91:59-69. - PubMed
-
- Bell, S. P. 2002. The origin recognition complex: from simple origins to complex functions. Genes Dev. 16:659-672. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
