Role of CTCF binding sites in the Igf2/H19 imprinting control region
- PMID: 15143173
- PMCID: PMC416431
- DOI: 10.1128/MCB.24.11.4791-4800.2004
Role of CTCF binding sites in the Igf2/H19 imprinting control region
Abstract
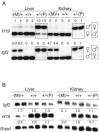
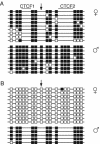
A approximately 2.4-kb imprinting control region (ICR) regulates somatic monoallelic expression of the Igf2 and H19 genes. This is achieved through DNA methylation-dependent chromatin insulator and promoter silencing activities on the maternal and paternal chromosomes, respectively. In somatic cells, the hypomethylated maternally inherited ICR binds the insulator protein CTCF at four sites and blocks activity of the proximal Igf2 promoter by insulating it from its distal enhancers. CTCF binding is thought to play a direct role in inhibiting methylation of the ICR in female germ cells and in somatic cells and, therefore, in establishing and maintaining imprinting of the Igf2/H19 region. Here, we report on the effects of eliminating ICR CTCF binding by severely mutating all four sites in mice. We found that in the female and male germ lines, the mutant ICR remained hypomethylated and hypermethylated, respectively, showing that the CTCF binding sites are dispensable for imprinting establishment. Postfertilization, the maternal mutant ICR acquired methylation, which could be explained by loss of methylation inhibition, which is normally provided by CTCF binding. Adjacent regions in cis-the H19 promoter and gene-also acquired methylation, accompanied by downregulation of H19. This could be the result of a silencing effect of the methylated maternal ICR.
Figures






References
-
- Bartolomei, M. S., and S. M. Tilghman. 1997. Genomic imprinting in mammals. Annu. Rev. Genet. 31:493-525. - PubMed
-
- Bartolomei, M. S., A. L. Webber, M. E. Brunkow, and S. M. Tilghman. 1993. Epigenetic mechanisms underlying the imprinting of the mouse H19 gene. Genes Dev. 7:1663-1673. - PubMed
-
- Bell, A. C., and G. Felsenfeld. 2000. Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature 405:482-485. - PubMed
-
- Bird, A. 2002. DNA methylation patterns and epigenetic memory. Genes Dev. 16:6-21. - PubMed
-
- Brandeis, M., D. Frank, I. Keshet, Z. Siegfried, M. Mendelsohn, A. Nemes, V. Temper, A. Razin, and H. Cedar. 1994. Sp1 elements protect a CpG island from de novo methylation. Nature 371:435-438. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
