Abl interactor 1 (Abi-1) wave-binding and SNARE domains regulate its nucleocytoplasmic shuttling, lamellipodium localization, and wave-1 levels
- PMID: 15143189
- PMCID: PMC416433
- DOI: 10.1128/MCB.24.11.4979-4993.2004
Abl interactor 1 (Abi-1) wave-binding and SNARE domains regulate its nucleocytoplasmic shuttling, lamellipodium localization, and wave-1 levels
Abstract
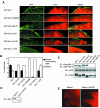
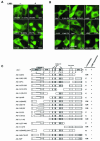
The Abl interactor 1 (Abi-1) protein has been implicated in the regulation of actin dynamics and localizes to the tips of lamellipodia and filopodia. Here, we show that Abi-1 binds the actin nucleator protein Wave-1 through an amino-terminal Wave-binding (WAB) domain and that disruption of the Abi-1-Wave-1 interaction prevents Abi-1 from reaching the tip of the lamellipodium. Abi-1 binds to the Wave homology domain of Wave-1, a region that is required for translocation of Wave-1 to the lamellipodium. Mouse embryo fibroblasts that lack one allele of Abi-1 and are homozygous null for the related Abi-2 protein exhibit decreased Wave-1 protein levels. This phenotype is rescued by Abi-1 proteins that retain Wave-1 binding but not by Abi-1 mutants that cannot bind to Wave-1. Moreover, we uncovered an overlapping SNARE domain in the amino terminus of Abi-1 that interacts with Syntaxin-1, a SNARE family member. Further, we demonstrated that Abi-1 shuttles in and out of the nucleus in a leptomycin B (LMB)-dependent manner and that complete nuclear translocation of Abi-1 in the absence of LMB requires the combined inactivation of the SNARE, WAB, and SH3 domains of Abi-1. Thus, Abi-1 undergoes nucleocytoplasmic shuttling and functions at the leading edge to regulate Wave-1 localization and protein levels.
Figures






References
-
- Beites, C. L., H. Xie, R. Bowser, and W. S. Trimble. 1999. The septin CDCrel-1 binds syntaxin and inhibits exocytosis. Nat. Neurosci. 5:434-439. - PubMed
-
- Biesova, Z., C. Piccoli, and W. T. Wong. 1997. Isolation and characterization of e3B1, an eps8 binding protein that regulates cell growth. Oncogene 14:233-241. - PubMed
-
- Blagg, S. L., M. Stewart, C. Sambles, and R. H. Insall. 2003. PIR121 regulates pseudopod dynamics and SCAR activity in Dictyostelium. Curr. Biol. 13:1480-1487. - PubMed
-
- Chapman, E. R., S. An, N. Barton, and R. Jahn. 1994. SNAP-25, a t-SNARE which binds to both syntaxin and synaptobrevin via domains that may form coiled coils. J. Biol. Chem. 269:27427-27432. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
