Nuclear surveillance and degradation of hypomodified initiator tRNAMet in S. cerevisiae
- PMID: 15145828
- PMCID: PMC420349
- DOI: 10.1101/gad.1183804
Nuclear surveillance and degradation of hypomodified initiator tRNAMet in S. cerevisiae
Abstract
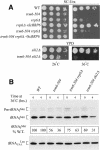
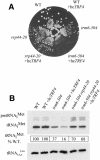
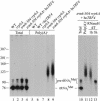
The tRNA m(1)A58 methyltransferase is composed of two subunits encoded by the essential genes TRM6 and TRM61 (formerly GCD10 and GCD14). The trm6-504 mutation results in a defective m(1)A methyltransferase (Mtase) and a temperature-sensitive growth phenotype that is attributable to the absence of m(1)A58 and consequential tRNA(i)(Met) instability. We used a genetic approach to identify the genes responsible for tRNA(i)(Met) degradation in trm6 cells. Three recessive extragenic mutations that suppress trm6-504 mutant phenotypes and restore hypomodified tRNA(i)(Met) to near normal levels were identified. The wild-type allele of one suppressor, DIS3/RRP44, encodes a 3'-5' exoribonuclease and a member of the multisubunit exosome complex. We provide evidence that a functional nuclear exosome is required for the degradation of tRNA(i)(Met) lacking m(1)A58. A second suppressor gene encodes Trf4p, a DNA polymerase (pol sigma) with poly(A) polymerase activity. Whereas deletion of TRF4 leads to stabilization of tRNA(i)(Met), overexpression of Trf4p destabilizes the hypomodified tRNA(i)(Met) in trm6 cells. The hypomodified, but not wild-type, pre-tRNA(i)(Met) accumulates as a polyadenylated species, whose abundance and length distribution both increase upon Trf4p overexpression. These data indicate that a tRNA surveillance pathway exists in yeast that requires Trf4p and the exosome for polyadenylation and degradation of hypomodified pre-tRNA(i)(Met).
Figures




References
-
- Anderson, J., Phan, L., Cuesta, R., Carlson, B.A., Pak, M., Asano, K., Bjork, G.R., Tamame, M., and Hinnebusch, A.G. 1998. The essential Gcd10p-Gcd14p nuclear complex is required for 1-methyladenosine modification and maturation of initiator methionyl-tRNA. Genes & Dev. 12: 3650-3662. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
