The entire Nup107-160 complex, including three new members, is targeted as one entity to kinetochores in mitosis
- PMID: 15146057
- PMCID: PMC452587
- DOI: 10.1091/mbc.e03-12-0878
The entire Nup107-160 complex, including three new members, is targeted as one entity to kinetochores in mitosis
Abstract
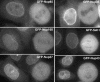
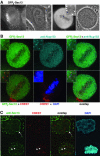
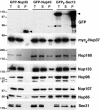
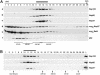
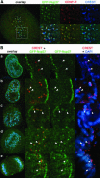
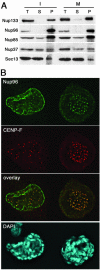
In eukaryotes, bidirectional transport of macromolecules between the cytoplasm and the nucleus occurs through elaborate supramolecular structures embedded in the nuclear envelope, the nuclear pore complexes (NPCs). NPCs are composed of multiple copies of approximately 30 different proteins termed nucleoporins, of which several can be biochemically isolated as subcomplexes. One such building block of the NPC, termed the Nup107-160 complex in vertebrates, was so far demonstrated to be composed of six different nucleoporins. Here, we identify three WD (Trp-Asp)-repeat nucleoporins as new members of this complex, two of which, Nup37 and Nup43, are specific to higher eukaryotes. The third new member Seh1 is more loosely associated with the Nup107-160 complex biochemically, but its depletion by RNA interference leads to phenotypes similar to knock down of other constituents of this complex. By combining green fluorescent protein-tagged nucleoporins and specific antibodies, we show that all the constituents of this complex, including Nup37, Nup43, Seh1, and Sec13, are targeted to kinetochores from prophase to anaphase of mitosis. Together, our results indicate that the entire Nup107-160 complex, which comprises nearly one-third of the so-far identified nucleoporins, specifically localizes to kinetochores in mitosis.
Figures


References
-
- Allen, N.P.C., Patel, S.S., Huang, L., Chalkley, R.J., Burlingame, A., Lutzmann, M., Hurt, E.C., and Rexach, M. (2002). Deciphering networks of protein interactions at the nuclear pore complex. Mol. Cell. Proteomics 1, 930-946. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

