Membrane lipid biosynthesis in Chlamydomonas reinhardtii: expression and characterization of CTP:phosphoethanolamine cytidylyltransferase
- PMID: 15147238
- PMCID: PMC1133914
- DOI: 10.1042/BJ20040254
Membrane lipid biosynthesis in Chlamydomonas reinhardtii: expression and characterization of CTP:phosphoethanolamine cytidylyltransferase
Abstract
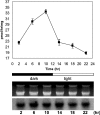
CTP:phosphoethanolamine cytidylyltransferase (ECT) is considered to be the regulatory enzyme in the CDP-ethanolamine pathway of phosphatidylethanolamine (PE) biosynthesis. The ECT cDNA of Chlamydomonas reinhardtii encodes a protein of 443 amino acid residues, which is longer than the same protein in yeast, rat or human. The translated product of cloned cDNA was expressed as a fusion protein in Escherichia coli, and was shown to have ECT activity. The deduced amino acid sequence has 41% identity with that of human or rat, and 30% with yeast. The ECT protein has a repetitive internal sequence in its N- and C-terminal halves and a signature peptide sequence, RTXGVSTT, typical of the cytidylyltransferase family. The first 70 amino acid residues do not match the N-terminal part of the cytidylyltransferases from other organisms, and we hypothesize that it is a subcellular targeting signal to mitochondria. ECT and organelle marker enzyme assays showed that the total activity of ECT correlates well with that of fumarase, a marker enzyme for mitochondria. Northern blots showed an increase in mRNA abundance during reflagellation, indicating a possibility of transcriptional regulation. A notable change in the enzyme activity in C. reinhardtii cells was observed during the cell cycle, increasing during the dark and then decreasing during the light period, while the mRNA level did not alter, providing evidence for post-translational regulation.
Figures


Similar articles
-
Cloning and expression of CTP:phosphoethanolamine cytidylyltransferase cDNA from rat liver.Biochem J. 1999 Oct 1;343 Pt 1(Pt 1):107-14. Biochem J. 1999. PMID: 10493918 Free PMC article.
-
Cloning of a human cDNA for CTP-phosphoethanolamine cytidylyltransferase by complementation in vivo of a yeast mutant.J Biol Chem. 1997 Apr 4;272(14):9567-72. doi: 10.1074/jbc.272.14.9567. J Biol Chem. 1997. PMID: 9083101
-
Isolation and characterization of ECT1 gene encoding CTP: phosphoethanolamine cytidylyltransferase of Saccharomyces cerevisiae.J Biochem. 1996 Nov;120(5):1040-7. doi: 10.1093/oxfordjournals.jbchem.a021497. J Biochem. 1996. PMID: 8982874
-
CTP:phosphoethanolamine cytidylyltransferase.Biochim Biophys Acta. 1997 Sep 4;1348(1-2):91-9. doi: 10.1016/s0005-2760(97)00113-6. Biochim Biophys Acta. 1997. PMID: 9370320 Review.
-
Metabolic and molecular aspects of ethanolamine phospholipid biosynthesis: the role of CTP:phosphoethanolamine cytidylyltransferase (Pcyt2).Biochem Cell Biol. 2007 Jun;85(3):283-300. doi: 10.1139/o07-006. Biochem Cell Biol. 2007. PMID: 17612623 Review.
Cited by
-
FT genome A and D polymorphisms are associated with the variation of earliness components in hexaploid wheat.Theor Appl Genet. 2008 Feb;116(3):383-94. doi: 10.1007/s00122-007-0676-0. Epub 2007 Nov 27. Theor Appl Genet. 2008. PMID: 18040656
-
Functional characterization of the Chlamydomonas reinhardtii ERG3 ortholog, a gene involved in the biosynthesis of ergosterol.PLoS One. 2010 Jan 11;5(1):e8659. doi: 10.1371/journal.pone.0008659. PLoS One. 2010. PMID: 20084111 Free PMC article.
-
Annotation of genes involved in glycerolipid biosynthesis in Chlamydomonas reinhardtii: discovery of the betaine lipid synthase BTA1Cr.Eukaryot Cell. 2005 Feb;4(2):242-52. doi: 10.1128/EC.4.2.242-252.2005. Eukaryot Cell. 2005. PMID: 15701786 Free PMC article.
-
The ethanolamine branch of the Kennedy pathway is essential in the bloodstream form of Trypanosoma brucei.Mol Microbiol. 2009 Sep;73(5):826-43. doi: 10.1111/j.1365-2958.2009.06764.x. Epub 2009 Jun 23. Mol Microbiol. 2009. PMID: 19555461 Free PMC article.
-
Defects in CTP:PHOSPHORYLETHANOLAMINE CYTIDYLYLTRANSFERASE affect embryonic and postembryonic development in Arabidopsis.Plant Cell. 2006 Dec;18(12):3370-85. doi: 10.1105/tpc.106.040840. Epub 2006 Dec 22. Plant Cell. 2006. PMID: 17189343 Free PMC article.
References
-
- Kent C. Eukaryotic phospholipid biosynthesis. Annu. Rev. Biochem. 1995;64:315–343. - PubMed
-
- Moore T. S. Enzymes of phospholipids synthesis. Methods in Plant Biochemistry, Enzymes of Primary Metabolism, vol. 3. In: Lea P. J., editor. London: Academic Press; 1990. pp. 229–239.
-
- Sundler R., Akesson B. Regulation of phospholipid biosynthesis in isolated rat hepatocytes: effect of different substrates. J. Biol. Chem. 1975;250:3359–3367. - PubMed
-
- Min-Seok R., Kawamata Y., Nakamura H., Ohta A., Takagi M. Isolation and characterization of ECT1 gene encoding CTP:phosphoethanolamine cytidylyltransferase of Saccharomyces cerevisiae. J. Biochem. (Tokyo) 1996;120:1040–1047. - PubMed
-
- Nakashima A., Hosaka K., Nikawa J. I. Cloning of a human cDNA for CTP-phosphoethanolamine cytidylyltransferase by complementation in vivo of a yeast mutant. J. Biol. Chem. 1997;272:9567–9572. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

