Characterization of SARS main protease and inhibitor assay using a fluorogenic substrate
- PMID: 15147951
- PMCID: PMC7134607
- DOI: 10.1016/j.bbrc.2004.04.098
Characterization of SARS main protease and inhibitor assay using a fluorogenic substrate
Erratum in
- Biochem Biophys Res Commun. 2004 Jul 23;320(2):623
Abstract
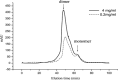
SARS main protease is essential for life cycle of SARS coronavirus and may be a key target for developing anti-SARS drugs. Recently, the enzyme expressed in Escherichia coli was characterized using a HPLC assay to monitor the formation of products from 11 peptide substrates covering the cleavage sites found in the SARS viral genome. This protease easily dissociated into inactive monomer and the deduced Kd of the dimer was 100 microM. In order to detect enzyme activity, the assay needed to be performed at micromolar enzyme concentration. This makes finding the tight inhibitor (nanomolar range IC50) impossible. In this study, we prepared a peptide with fluorescence quenching pair (Dabcyl and Edans) at both ends of a peptide substrate and used this fluorogenic peptide substrate to characterize SARS main protease and screen inhibitors. The fluorogenic peptide gave extremely sensitive signal upon cleavage catalyzed by the protease. Using this substrate, the protease exhibits a significantly higher activity (kcat = 1.9 s(-1) and Km = 17 microM) compared to the previously reported parameters. Under our assay condition, the enzyme stays as an active dimer without dissociating into monomer and reveals a small Kd value (15 nM). This enzyme in conjunction with fluorogenic peptide substrate provides us a suitable tool for identifying potent inhibitors of SARS protease.
Figures



Similar articles
-
A continuous fluorescence-based assay of human cytomegalovirus protease using a peptide substrate.Anal Biochem. 1995 May 1;227(1):148-55. doi: 10.1006/abio.1995.1264. Anal Biochem. 1995. PMID: 7668375
-
Novel fluorogenic substrates for assaying retroviral proteases by resonance energy transfer.Science. 1990 Feb 23;247(4945):954-8. doi: 10.1126/science.2106161. Science. 1990. PMID: 2106161
-
Characterization of SARS-CoV main protease and identification of biologically active small molecule inhibitors using a continuous fluorescence-based assay.FEBS Lett. 2004 Oct 22;576(3):325-30. doi: 10.1016/j.febslet.2004.09.026. FEBS Lett. 2004. PMID: 15498556 Free PMC article.
-
Characterization and inhibition of SARS-coronavirus main protease.Curr Top Med Chem. 2006;6(4):361-76. doi: 10.2174/156802606776287090. Curr Top Med Chem. 2006. PMID: 16611148 Review.
-
Quaternary structure, substrate selectivity and inhibitor design for SARS 3C-like proteinase.Curr Pharm Des. 2006;12(35):4555-64. doi: 10.2174/138161206779010396. Curr Pharm Des. 2006. PMID: 17168761 Review.
Cited by
-
Mechanism of the maturation process of SARS-CoV 3CL protease.J Biol Chem. 2005 Sep 2;280(35):31257-66. doi: 10.1074/jbc.M502577200. Epub 2005 Mar 23. J Biol Chem. 2005. PMID: 15788388 Free PMC article.
-
Dynamically-driven inactivation of the catalytic machinery of the SARS 3C-like protease by the N214A mutation on the extra domain.PLoS Comput Biol. 2011 Feb;7(2):e1001084. doi: 10.1371/journal.pcbi.1001084. Epub 2011 Feb 24. PLoS Comput Biol. 2011. PMID: 21390281 Free PMC article.
-
Efficiency Improvements and Discovery of New Substrates for a SARS-CoV-2 Main Protease FRET Assay.SLAS Discov. 2021 Oct;26(9):1189-1199. doi: 10.1177/24725552211020681. Epub 2021 Jun 19. SLAS Discov. 2021. PMID: 34151620 Free PMC article.
-
DNA-encoded chemistry technology yields expedient access to SARS-CoV-2 Mpro inhibitors.Proc Natl Acad Sci U S A. 2021 Sep 7;118(36):e2111172118. doi: 10.1073/pnas.2111172118. Proc Natl Acad Sci U S A. 2021. PMID: 34426525 Free PMC article.
-
Mutation of Asn28 disrupts the dimerization and enzymatic activity of SARS 3CL(pro).Biochemistry. 2010 May 25;49(20):4308-17. doi: 10.1021/bi1002585. Biochemistry. 2010. PMID: 20420403 Free PMC article.
References
-
- Drosten C., Gunther S., Preiser W., van der Werf S., Brodt H.R., Becker S., Rabenau H., Panning M., Kolesnikova L., Fouchier R.A., Berger A., Burguiere A.M., Cinatl J., Eickmann M., Escriou N., Grywna K., Kramme S., Manuguerra J.C., Muller S., Rickerts V., Sturmer M., Vieth S., Klenk H.D., Osterhaus A.D., Schmitz H., Doerr H.W. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348:1967–1976. - PubMed
-
- Ksiazek T.G., Erdman D., Golsmith C.S., Zaki S.R., Peret T., Emery S., Tong S., Urbani C., Comer J.A., Lim W., Rollin P.E., Dowell S.F., Ling A.E., Humphrey C.D., Shieh W.J., Guarner J., Paddock C.D., Rota P., Fields B., DeRisis J., Yang J.Y., Cox N., Hughes J.M., LeDue J.W., Bellini W.J., Anderson L.J.A. A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348:1953–1966. - PubMed
-
- Rota P.A., Oberste M.S., Monroe S.S., Nix W.A., Campagnoli R., Icenogle J.P., Penaranda S., Bankamp B., Maher K., Chen M.-H., Tong S., Tamin A., Lowe L., Frace M., Derisi J.L., Chen Q., Wang D., Erdman D.D., Peret T.C.T., Burns C., Ksiazek T.G., Rollin P.E., Sanchez A., Liffick S., Holloway B., Limor J., McCaustland K., Olsen-Rasmussen M., Fouchier R., Gunther S., Osterhaus A.D.M.E., Drosten C., Pallansch M.A., Anderson L.J., Bellini W.J. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science. 2003;300:1394–1399. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

