Endogenous presentation of CD8+ T cell epitopes from Epstein-Barr virus-encoded nuclear antigen 1
- PMID: 15148340
- PMCID: PMC2211806
- DOI: 10.1084/jem.20040191
Endogenous presentation of CD8+ T cell epitopes from Epstein-Barr virus-encoded nuclear antigen 1
Abstract
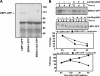
Epstein-Barr virus (EBV)-encoded nuclear antigen (EBNA)1 is thought to escape cytotoxic T lymphocyte (CTL) recognition through either self-inhibition of synthesis or by blockade of proteasomal degradation by the glycine-alanine repeat (GAr) domain. Here we show that EBNA1 has a remarkably varied cell type-dependent stability. However, these different degradation rates do not correspond to the level of major histocompatibility complex class I-restricted presentation of EBNA1 epitopes. In spite of the highly stable expression of EBNA1 in B cells, CTL epitopes derived from this protein are efficiently processed and presented to CD8+ T cells. Furthermore, we show that EBV-infected B cells can readily activate EBNA1-specific memory T cell responses from healthy virus carriers. Functional assays revealed that processing of these EBNA1 epitopes is proteasome and transporter associated with antigen processing dependent. We also show that the endogenous presentation of these epitopes is dependent on the newly synthesized protein rather than the long-lived stable EBNA1. Based on these observations, we propose that defective ribosomal products, not the full-length antigen, are the primary source of endogenously processed CD8+ T cell epitopes from EBNA1.
Figures




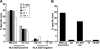
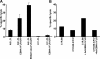
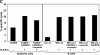
Similar articles
-
CD8 T cell recognition of endogenously expressed epstein-barr virus nuclear antigen 1.J Exp Med. 2004 May 17;199(10):1409-20. doi: 10.1084/jem.20040121. J Exp Med. 2004. PMID: 15148339 Free PMC article.
-
Self-inhibition of synthesis and antigen presentation by Epstein-Barr virus-encoded EBNA1.Science. 2003 Sep 5;301(5638):1371-4. doi: 10.1126/science.1088902. Science. 2003. PMID: 12958359
-
Full-length EBNA1 mRNA-transduced dendritic cells stimulate cytotoxic T lymphocytes recognizing a novel HLA-Cw*0303- and -Cw*0304-restricted epitope on EBNA1-expressing cells.J Gen Virol. 2007 Mar;88(Pt 3):770-780. doi: 10.1099/vir.0.82519-0. J Gen Virol. 2007. PMID: 17325349
-
Epstein-Barr virus evasion of CD8(+) and CD4(+) T cell immunity via concerted actions of multiple gene products.Semin Cancer Biol. 2008 Dec;18(6):397-408. doi: 10.1016/j.semcancer.2008.10.008. Epub 2008 Oct 25. Semin Cancer Biol. 2008. PMID: 18977445 Review.
-
Epstein-Barr virus-encoded EBNA1 and ZEBRA: targets for therapeutic strategies against EBV-carrying cancers.J Pathol. 2015 Jan;235(2):334-41. doi: 10.1002/path.4431. J Pathol. 2015. PMID: 25186125 Review.
Cited by
-
Characterization of an human leucocyte antigen A2-restricted Epstein-Barr virus nuclear antigen-1-derived cytotoxic T-lymphocyte epitope.Immunology. 2010 Mar;129(3):386-95. doi: 10.1111/j.1365-2567.2009.03190.x. Epub 2009 Nov 16. Immunology. 2010. PMID: 19922423 Free PMC article.
-
Immunotherapy for EBV-associated malignancies.Int J Hematol. 2011 Mar;93(3):281-293. doi: 10.1007/s12185-011-0782-2. Epub 2011 Feb 19. Int J Hematol. 2011. PMID: 21336546
-
mRNA Structural constraints on EBNA1 synthesis impact on in vivo antigen presentation and early priming of CD8+ T cells.PLoS Pathog. 2014 Oct 9;10(10):e1004423. doi: 10.1371/journal.ppat.1004423. eCollection 2014 Oct. PLoS Pathog. 2014. PMID: 25299404 Free PMC article.
-
Therapeutic implications of Epstein-Barr virus infection for the treatment of nasopharyngeal carcinoma.Ther Clin Risk Manag. 2014 Sep 5;10:721-36. doi: 10.2147/TCRM.S47434. eCollection 2014. Ther Clin Risk Manag. 2014. PMID: 25228810 Free PMC article. Review.
-
Limited Recognition of Highly Conserved Regions of SARS-CoV-2.Microbiol Spectr. 2022 Feb 23;10(1):e0278021. doi: 10.1128/spectrum.02780-21. Epub 2022 Feb 23. Microbiol Spectr. 2022. PMID: 35196796 Free PMC article.
References
-
- Benoist, C., and D. Mathis. 1992. Generation of the alpha beta T-cell repertoire. Curr. Opin. Immunol. 4:156–161. - PubMed
-
- Khanna, R., and S.R. Burrows. 2000. Role of cytotoxic T lymphocytes in Epstein-Barr virus-associated diseases. Annu. Rev. Microbiol. 54:19–48. - PubMed
-
- Tortorella, D., B.E. Gewurz, M.H. Furman, D.J. Schust, and H.L. Ploegh. 2000. Viral subversion of the immune system. Annu. Rev. Immunol. 18:861–926. - PubMed
-
- Murray, R.J., M.G. Kurilla, J.M. Brooks, W.A. Thomas, M. Rowe, E. Kieff, and A.B. Rickinson. 1992. Identification of target antigens for the human cytotoxic T-cell response to Epstein-Barr virus (EBV): implications for the immune control of EBV-positive malignancies. J. Exp. Med. 176:157–168. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

