CsdA, a cold-shock RNA helicase from Escherichia coli, is involved in the biogenesis of 50S ribosomal subunit
- PMID: 15148362
- PMCID: PMC419605
- DOI: 10.1093/nar/gkh603
CsdA, a cold-shock RNA helicase from Escherichia coli, is involved in the biogenesis of 50S ribosomal subunit
Abstract
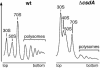
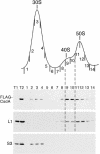
CsdA, a DEAD-box protein from Escherichia coli, has been proposed to participate in a variety of processes, such as translation initiation, gene regulation after cold-shock, mRNA decay and biogenesis of the small ribosomal subunit. Whether the protein really plays a direct role in these multiple processes is however, not clear. Here, we show that CsdA is involved in the biogenesis of the large rather than the small ribosomal subunit. Deletion of the csdA gene leads to a deficit in free 50S subunits at low temperatures and to the accumulation of a new particle sedimenting around 40S. Analysis of the RNA and protein contents of this particle indicates that it corresponds to a mis-assembled large subunit. Sucrose gradient fractionation shows that in wild-type cells CsdA associates mainly with a pre50S particle. Presumably the RNA helicase activity of CsdA permits a structural rearrangement during 50S biogenesis at low temperature. We showed previously that SrmB, another DEAD-box RNA helicase, is also involved in 50S assembly in E.coli. Our results suggest that CsdA is required at a later step than SrmB. However, over-expression of CsdA corrects the ribosome defect of the srmB-deleted strain, indicating that some functional overlap exists between the two proteins.
Figures





References
-
- Linder P., Lasko,P.F., Ashburner,M., Leroy,P., Nielsen,P.J., Nishi,K., Schnier,J. and Slonimski,P.P. (1989) Birth of the DEAD box. Nature, 337, 121–122. - PubMed
-
- Tanner N.K. and Linder,P. (2001) DExD/H box RNA helicases: from generic motors to specific dissociation functions. Mol. Cell., 8, 251–262. - PubMed
-
- de la Cruz J., Kressler,D. and Linder,P. (1999) Unwinding RNA in Saccharomyces cerevisiae: DEAD-box proteins and related families. Trends Biochem. Sci., 24, 192–198. - PubMed
-
- Lorsch J.R. (2002) RNA chaperones exist and DEAD box proteins get a life. Cell, 109, 797–800. - PubMed
-
- Linder P., Tanner,N.K. and Banroques,J. (2001) From RNA helicases to RNPases. Trends Biochem. Sci., 26, 339–341. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

