Geometry and symmetry presculpt the free-energy landscape of proteins
- PMID: 15148372
- PMCID: PMC419539
- DOI: 10.1073/pnas.0402525101
Geometry and symmetry presculpt the free-energy landscape of proteins
Abstract
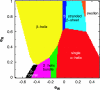
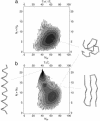
We present a simple physical model that demonstrates that the native-state folds of proteins can emerge on the basis of considerations of geometry and symmetry. We show that the inherent anisotropy of a chain molecule, the geometrical and energetic constraints placed by the hydrogen bonds and sterics, and hydrophobicity are sufficient to yield a free-energy landscape with broad minima even for a homopolymer. These minima correspond to marginally compact structures comprising the menu of folds that proteins choose from to house their native states in. Our results provide a general framework for understanding the common characteristics of globular proteins.
Figures

Similar articles
-
Geometrical model for the native-state folds of proteins.Biophys Chem. 2005 Apr 1;115(2-3):289-94. doi: 10.1016/j.bpc.2004.12.036. Epub 2004 Dec 24. Biophys Chem. 2005. PMID: 15752620
-
Importance of metastable states in the free energy landscapes of polypeptide chains.Phys Rev Lett. 2007 Oct 26;99(17):178104. doi: 10.1103/PhysRevLett.99.178104. Epub 2007 Oct 26. Phys Rev Lett. 2007. PMID: 17995375
-
II. Geometrical framework for thinking about globular proteins: The power of poking.Proteins. 2025 Jan;93(1):160-175. doi: 10.1002/prot.26566. Epub 2023 Aug 21. Proteins. 2025. PMID: 37605446
-
Water as a Good Solvent for Unfolded Proteins: Folding and Collapse are Fundamentally Different.J Mol Biol. 2020 Apr 17;432(9):2882-2889. doi: 10.1016/j.jmb.2020.01.031. Epub 2020 Feb 7. J Mol Biol. 2020. PMID: 32044346 Free PMC article. Review.
-
How hydrogen bonds shape membrane protein structure.Adv Protein Chem. 2005;72:157-72. doi: 10.1016/S0065-3233(05)72006-4. Adv Protein Chem. 2005. PMID: 16581376 Review.
Cited by
-
A glimpse at the organization of the protein universe.Proc Natl Acad Sci U S A. 2005 Apr 19;102(16):5641-2. doi: 10.1073/pnas.0500274102. Epub 2005 Apr 12. Proc Natl Acad Sci U S A. 2005. PMID: 15827120 Free PMC article. No abstract available.
-
Toward Consistent Physics-Based Modeling of Local Backbone Structures and Chirality Change of Proteins in Coarse-Grained Approaches.J Phys Chem Lett. 2023 Nov 9;14(44):9824-9833. doi: 10.1021/acs.jpclett.3c01988. Epub 2023 Oct 27. J Phys Chem Lett. 2023. PMID: 37889895 Free PMC article. Review.
-
Mapping the Protein Fold Universe Using the CamTube Force Field in Molecular Dynamics Simulations.PLoS Comput Biol. 2015 Oct 27;11(10):e1004435. doi: 10.1371/journal.pcbi.1004435. eCollection 2015 Oct. PLoS Comput Biol. 2015. PMID: 26505754 Free PMC article.
-
The Iconic α-Helix: From Pauling to the Present.Methods Mol Biol. 2025;2867:1-17. doi: 10.1007/978-1-0716-4196-5_1. Methods Mol Biol. 2025. PMID: 39576572
-
Secondary Structure in Free and Assisted Modeling of Proteins with the Coarse-Grained UNRES Force Field.Methods Mol Biol. 2025;2867:19-41. doi: 10.1007/978-1-0716-4196-5_2. Methods Mol Biol. 2025. PMID: 39576573
References
-
- Wolynes, P. G., Onuchic, J. N. & Thirumalai, D. (1995) Science 267, 1619–1620. - PubMed
-
- Dill, K. A. & Chan, H. S. (1997) Nat. Struct. Biol. 4, 10–19. - PubMed
-
- Dobson, C. M., Sali, A. & Karplus, M. (1998) Angew. Chem. Int. Ed. Engl. 37, 868–893. - PubMed
-
- Fersht, A. R. (1998) Structure and Mechanism in Protein Science: A Guide to Enzyme Catalysis and Protein Folding (Freeman, New York).
-
- Baker, D. (2002) Nature 405, 39–42. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

