XLalphas, the extra-long form of the alpha-subunit of the Gs G protein, is significantly longer than suspected, and so is its companion Alex
- PMID: 15148396
- PMCID: PMC420400
- DOI: 10.1073/pnas.0308758101
XLalphas, the extra-long form of the alpha-subunit of the Gs G protein, is significantly longer than suspected, and so is its companion Alex
Abstract
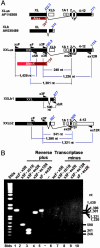
Because of the use of alternate exons 1, mammals express two distinct forms of Gsalpha-subunits: the canonical 394-aa Gsalpha present in all tissues and a 700+-aa extra-long alphas (XLalphas) expressed in a more restricted manner. Both subunits transduce receptor signals into stimulation of adenylyl cyclase. The XL exon encodes the XL domain of XLalphas and, in a parallel ORF, a protein called Alex. Alex interacts with the XL domain of XLalphas and inhibits its adenylyl cyclase-stimulating function. In mice, rats, and humans, the XL exon is thought to contribute 422.3, 367.3, and 551.3 codons and to encode Alex proteins of 390, 357, and 561 aa, respectively. We report here that the XL exon is longer than presumed and contributes in mice, rats, and humans, respectively, an additional 364, 430, and 139 codons to XLalphas. We called the N-terminally extended XLalphas extra-extra-long Gsalpha, or XXLalphas. Alex is likewise longer. Its ORF also remains open in the 5' direction for approximately 2,000 nt, giving rise to Alex-extended, or AlexX. RT-PCR of murine total brain RNA shows that the entire XXL domain is encoded in a single exon. Furthermore, we discovered two truncated forms of XXLalphas, XXLb1 and XXLb2, in which, because of alternative splicing, the Gsalpha domain is replaced by different sequences. XXLb proteins are likely to be found as stable dimers with AlexX. The N-terminally longer proteins may play regulatory roles.
Figures





References
-
- Weinstein, L. S. (2001) J. Clin. Endocrinol. Metab. 86, 4622-4626. - PubMed
-
- Ischia, R., Lovisetti-Scamihorn, P., Hogue-Angeletti, R., Wolkersdorfer, M., Winkler, H. & Fischer-Colbrie, R. (1997) J. Biol. Chem. 272, 11657-11662. - PubMed
-
- Kehlenbach, R. H., Matthey, J. & Huttner, W. B. (1994) Nature 372, 804-809. - PubMed
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

