The crystal structure of xanthine oxidoreductase during catalysis: implications for reaction mechanism and enzyme inhibition
- PMID: 15148401
- PMCID: PMC419534
- DOI: 10.1073/pnas.0400973101
The crystal structure of xanthine oxidoreductase during catalysis: implications for reaction mechanism and enzyme inhibition
Abstract
Molybdenum is widely distributed in biology and is usually found as a mononuclear metal center in the active sites of many enzymes catalyzing oxygen atom transfer. The molybdenum hydroxylases are distinct from other biological systems catalyzing hydroxylation reactions in that the oxygen atom incorporated into the product is derived from water rather than molecular oxygen. Here, we present the crystal structure of the key intermediate in the hydroxylation reaction of xanthine oxidoreductase with a slow substrate, in which the carbon-oxygen bond of the product is formed, yet the product remains complexed to the molybdenum. This intermediate displays a stable broad charge-transfer band at approximately 640 nm. The crystal structure of the complex indicates that the catalytically labile Mo-OH oxygen has formed a bond with a carbon atom of the substrate. In addition, the MoS group of the oxidized enzyme has become protonated to afford Mo-SH on reduction of the molybdenum center. In contrast to previous assignments, we find this last ligand at an equatorial position in the square-pyramidal metal coordination sphere, not the apical position. A water molecule usually seen in the active site of the enzyme is absent in the present structure, which probably accounts for the stability of this intermediate toward ligand displacement by hydroxide.
Figures


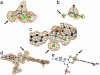



References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

