Uncommon deletions of the Smith-Magenis syndrome region can be recurrent when alternate low-copy repeats act as homologous recombination substrates
- PMID: 15148657
- PMCID: PMC1182010
- DOI: 10.1086/422016
Uncommon deletions of the Smith-Magenis syndrome region can be recurrent when alternate low-copy repeats act as homologous recombination substrates
Abstract
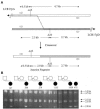
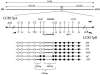
Several homologous recombination "hotspots," or sites of positional preference for strand exchanges, associated with recurrent deletions and duplications have been reported within large low-copy repeats (LCRs). Recently, such a hotspot was identified in patients with the Smith-Magenis syndrome (SMS) common deletion of approximately 4 Mb or a reciprocal duplication within the KER gene cluster of the SMS-REP LCRs, in which 50% of analyzed strand exchanges resulting in deletion and 23% of those resulting in duplication occurred. Here, we report an additional recombination hotspot within LCR17pA and LCR17pD, which serve as alternative substrates for nonallelic homologous recombination that results in large (approximately 5 Mb) deletions of 17p11.2, which include the SMS region. Using polymerase-chain-reaction mapping of somatic cell hybrid lines, we refined the breakpoints of six deletions within these LCRs. Sequence analysis of the recombinant junctions revealed that all six strand exchanges occurred within a 524-bp interval, and four of them occurred within an AluSq/x element. This interval represents only 0.5% of the 124-kb stretch of 98.6% sequence identity between LCR17pA and LCR17pD. A search for potentially stimulating sequence motifs revealed short AT-rich segments flanking the recombination hotspot. Our findings indicate that alternative LCRs can mediate rearrangements, resulting in haploinsufficiency of the SMS critical region, and reimplicate homologous recombination as a major mechanism for genomic disorders.
Figures
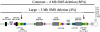
Similar articles
-
Reciprocal crossovers and a positional preference for strand exchange in recombination events resulting in deletion or duplication of chromosome 17p11.2.Am J Hum Genet. 2003 Dec;73(6):1302-15. doi: 10.1086/379979. Epub 2003 Nov 24. Am J Hum Genet. 2003. PMID: 14639526 Free PMC article.
-
Structure and evolution of the Smith-Magenis syndrome repeat gene clusters, SMS-REPs.Genome Res. 2002 May;12(5):729-38. doi: 10.1101/gr.82802. Genome Res. 2002. PMID: 11997339 Free PMC article.
-
Genetic proof of unequal meiotic crossovers in reciprocal deletion and duplication of 17p11.2.Am J Hum Genet. 2002 Nov;71(5):1072-81. doi: 10.1086/344346. Epub 2002 Oct 9. Am J Hum Genet. 2002. PMID: 12375235 Free PMC article.
-
Trisomy 17p10-p12 due to mosaic supernumerary marker chromosome: delineation of molecular breakpoints and clinical phenotype, and comparison to other proximal 17p segmental duplications.Am J Med Genet A. 2005 Oct 1;138A(2):175-80. doi: 10.1002/ajmg.a.30948. Am J Med Genet A. 2005. PMID: 16152635 Review.
-
New developments in Smith-Magenis syndrome (del 17p11.2).Curr Opin Neurol. 2007 Apr;20(2):125-34. doi: 10.1097/WCO.0b013e3280895dba. Curr Opin Neurol. 2007. PMID: 17351481 Review.
Cited by
-
Identification of a RAI1-associated disease network through integration of exome sequencing, transcriptomics, and 3D genomics.Genome Med. 2016 Nov 1;8(1):105. doi: 10.1186/s13073-016-0359-z. Genome Med. 2016. PMID: 27799067 Free PMC article.
-
Low copy repeats mediate distal chromosome 22q11.2 deletions: sequence analysis predicts breakpoint mechanisms.Genome Res. 2007 Apr;17(4):482-91. doi: 10.1101/gr.5986507. Epub 2007 Mar 9. Genome Res. 2007. PMID: 17351135 Free PMC article.
-
Complex chromosome 17p rearrangements associated with low-copy repeats in two patients with congenital anomalies.Hum Genet. 2007 Jul;121(6):697-709. doi: 10.1007/s00439-007-0359-6. Epub 2007 Apr 25. Hum Genet. 2007. PMID: 17457615 Free PMC article.
-
From microscopes to microarrays: dissecting recurrent chromosomal rearrangements.Nat Rev Genet. 2007 Nov;8(11):869-83. doi: 10.1038/nrg2136. Nat Rev Genet. 2007. PMID: 17943194 Free PMC article. Review.
-
Non-recurrent 17p11.2 deletions are generated by homologous and non-homologous mechanisms.Hum Genet. 2005 Jan;116(1-2):1-7. doi: 10.1007/s00439-004-1204-9. Epub 2004 Oct 22. Hum Genet. 2005. PMID: 15526218
References
Electronic-Database Information
-
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/
References
-
- Bi W, Yan J, Stankiewicz P, Park SS, Walz K, Boerkoel CF, Potocki L, Shaffer LG, Devriendt K, Nowaczyk MJ, Inoue K, Lupski JR (2002) Genes in a refined Smith-Magenis syndrome critical deletion interval on chromosome 17p11.2 and the syntenic region of the mouse. Genome Res 12:713–72810.1101/gr.73702 - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

