An accessory protein is required for relaxosome formation by small staphylococcal plasmids
- PMID: 15150221
- PMCID: PMC415746
- DOI: 10.1128/JB.186.11.3363-3373.2004
An accessory protein is required for relaxosome formation by small staphylococcal plasmids
Abstract
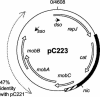
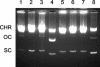
Mobilization of the staphylococcal plasmid pC221 requires at least one plasmid-encoded protein, MobA, in order to form a relaxosome. pC221 and closely related plasmids also possess an overlapping reading frame encoding a protein of 15 kDa, termed MobC. By completing the nucleotide sequence of plasmid pC223, we have found a further example of this small protein, and gene knockouts have shown that MobC is essential for relaxosome formation and plasmid mobilization in both pC221 and pC223. Primer extension analysis has been used to identify the nic site in both of these plasmids, located upstream of the mobC gene in the sense strand. Although the sequence surrounding the nic site is highly conserved between pC221 and pC223, exchange of the oriT sequence between plasmids significantly reduces the extent of relaxation complex formation, suggesting that the Mob proteins are selective for their cognate plasmids in vivo.
Figures

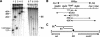


Similar articles
-
Investigating the basis of substrate recognition in the pC221 relaxosome.Mol Microbiol. 2006 Jun;60(5):1302-18. doi: 10.1111/j.1365-2958.2006.05188.x. Mol Microbiol. 2006. PMID: 16689804 Free PMC article.
-
Reconstitution of a staphylococcal plasmid-protein relaxation complex in vitro.J Bacteriol. 2004 Jun;186(11):3374-83. doi: 10.1128/JB.186.11.3374-3383.2004. J Bacteriol. 2004. PMID: 15150222 Free PMC article.
-
Two atypical mobilization proteins are involved in plasmid CloDF13 relaxation.Mol Microbiol. 2001 Feb;39(4):1088-99. doi: 10.1046/j.1365-2958.2001.02308.x. Mol Microbiol. 2001. PMID: 11251827
-
DNA processing reactions in bacterial conjugation.Annu Rev Biochem. 1995;64:141-69. doi: 10.1146/annurev.bi.64.070195.001041. Annu Rev Biochem. 1995. PMID: 7574478 Review.
-
Relaxosome function and conjugation regulation in F-like plasmids - a structural biology perspective.Mol Microbiol. 2012 Aug;85(4):602-17. doi: 10.1111/j.1365-2958.2012.08131.x. Epub 2012 Jul 13. Mol Microbiol. 2012. PMID: 22788760 Review.
Cited by
-
Genetic Dissection of a Prevalent Plasmid-Encoded Conjugation System in Lactococcus lactis.Front Microbiol. 2021 May 28;12:680920. doi: 10.3389/fmicb.2021.680920. eCollection 2021. Front Microbiol. 2021. PMID: 34122391 Free PMC article.
-
Mobilization and prevalence of a Fusobacterial plasmid.Plasmid. 2010 Jan;63(1):11-9. doi: 10.1016/j.plasmid.2009.09.001. Epub 2009 Sep 15. Plasmid. 2010. PMID: 19761790 Free PMC article.
-
Genomics of microbial plasmids: classification and identification based on replication and transfer systems and host taxonomy.Front Microbiol. 2015 Mar 31;6:242. doi: 10.3389/fmicb.2015.00242. eCollection 2015. Front Microbiol. 2015. PMID: 25873913 Free PMC article. Review.
-
Antimicrobial Resistance among Staphylococci of Animal Origin.Microbiol Spectr. 2018 Jul;6(4):10.1128/microbiolspec.arba-0010-2017. doi: 10.1128/microbiolspec.ARBA-0010-2017. Microbiol Spectr. 2018. PMID: 29992898 Free PMC article. Review.
-
Major families of multiresistant plasmids from geographically and epidemiologically diverse staphylococci.G3 (Bethesda). 2011 Dec;1(7):581-91. doi: 10.1534/g3.111.000760. Epub 2011 Dec 1. G3 (Bethesda). 2011. PMID: 22384369 Free PMC article.
References
-
- Apisiridej, S., A. Leelaporn, C. D. Scaramuzzi, R. A. Skurray, and N. Firth. 1997. Molecular analysis of a mobilizable theta-mode trimethoprim resistance plasmid from coagulase-negative staphylococci. Plasmid 38:13-24. - PubMed
-
- Aubert, S., K. G. Dyke, and N. E. Solh. 1998. Analysis of two Staphylococcus epidermidis plasmids coding for resistance to streptogramin A. Plasmid 40:238-242. - PubMed
-
- Augustin, J., and F. Gotz. 1990. Transformation of Staphylococcus epidermidis and other staphylococcal species with plasmid DNA by electroporation. FEMS Microbiol. Lett. 66:203-208. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

