Identification and characterization of glxR, a gene involved in regulation of glyoxylate bypass in Corynebacterium glutamicum
- PMID: 15150232
- PMCID: PMC415749
- DOI: 10.1128/JB.186.11.3453-3460.2004
Identification and characterization of glxR, a gene involved in regulation of glyoxylate bypass in Corynebacterium glutamicum
Abstract
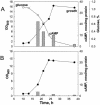
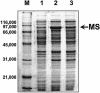
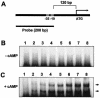
A corynebacterial clone, previously isolated by scoring repression of lacZYA fused to the aceB promoter of Corynebacterium glutamicum, was analyzed further. In the clone, an open reading frame designated glxR, consisting of 681 nucleotides and encoding a 24,957-Da protein, was found. The molecular mass of a native GlxR protein was estimated by gel filtration column chromatography to be 44,000 Da, suggesting that the protein formed dimers. The predicted amino acid sequence contained both cyclic AMP (cAMP)- and DNA-binding motifs and was homologous with the cAMP receptor protein family of proteins. The aceB-repressing activity of the glxR clone was markedly relieved in an Escherichia coli cya mutant, but the activity was restored in growth medium containing cAMP. In glucose medium, the intracellular cAMP concentration of C. glutamicum reached 22 nmol/mg of protein in the early exponential phase and then decreased further; but in acetate medium, the intracellular cAMP concentration was only 5 nmol/mg of protein and remained low throughout the growth phase. The expression of glxR was not affected by the carbon source. Binding of purified GlxR to the promoter region of aceB could be demonstrated only in the presence of cAMP. These data suggest that GlxR may form dimers which bind to the aceB promoter region in the presence of cAMP and repress the glyoxylate bypass genes.
Figures




References
-
- Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. - PubMed
-
- Bradford, M. M. 1976. A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. - PubMed
-
- Bruckner, R., and F. Titgemeyer. 2002. Carbon catabolite repression in bacteria: choice of the carbon source and autoregulatory limitation of sugar utilization. FEMS Microbiol. Lett. 209:141-148. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

