Novel genes that influence development in Streptomyces coelicolor
- PMID: 15150245
- PMCID: PMC415741
- DOI: 10.1128/JB.186.11.3570-3577.2004
Novel genes that influence development in Streptomyces coelicolor
Abstract
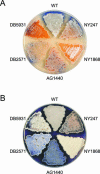
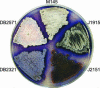
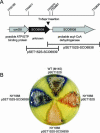
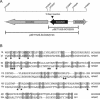
Filamentous soil bacteria of the genus Streptomyces carry out complex developmental cycles that result in sporulation and production of numerous secondary metabolites with pharmaceutically important activities. To further characterize the molecular basis of these developmental events, we screened for mutants of Streptomyces coelicolor that exhibit aberrant morphological differentiation and/or secondary metabolite production. On the basis of this screening analysis and the subsequent complementation analysis of the mutants obtained we assigned developmental roles to a gene involved in methionine biosynthesis (metH) and two previously uncharacterized genes (SCO6938 and SCO2525) and we reidentified two previously described developmental genes (bldA and bldM). In contrast to most previously studied genes involved in development, the genes newly identified in the present study all appear to encode biosynthetic enzymes instead of regulatory proteins. The MetH methionine synthase appears to be required for conversion of aerial hyphae into chains of spores, SCO6938 is a probable acyl coenzyme A dehydrogenase that contributes to the proper timing of aerial mycelium formation and antibiotic production, and SCO2525 is a putative methyltransferase that influences various aspects of colony growth and development.
Figures

Similar articles
-
rag genes: novel components of the RamR regulon that trigger morphological differentiation in Streptomyces coelicolor.Mol Microbiol. 2006 Sep;61(5):1167-86. doi: 10.1111/j.1365-2958.2006.05304.x. Mol Microbiol. 2006. PMID: 16925552
-
A novel Streptomyces gene, samR, with different effects on differentiation of Streptomyces ansochromogenes and Streptomyces coelicolor.Arch Microbiol. 2002 Mar;177(3):274-8. doi: 10.1007/s00203-001-0382-2. Epub 2002 Jan 8. Arch Microbiol. 2002. PMID: 11907684
-
The Leeuwenhoek lecture, 1987. Towards an understanding of gene switching in Streptomyces, the basis of sporulation and antibiotic production.Proc R Soc Lond B Biol Sci. 1988 Nov 22;235(1279):121-38. doi: 10.1098/rspb.1988.0067. Proc R Soc Lond B Biol Sci. 1988. PMID: 2907142 Review.
-
[Identification and functional characterization of sawE--a gene involved in differentiation of Streptomyces ansochromogenes 7100].Wei Sheng Wu Xue Bao. 2003 Feb;43(1):48-55. Wei Sheng Wu Xue Bao. 2003. PMID: 16276871 Chinese.
-
Regulation of Streptomyces development: reach for the sky!Trends Microbiol. 2006 Jul;14(7):313-9. doi: 10.1016/j.tim.2006.05.008. Epub 2006 Jun 6. Trends Microbiol. 2006. PMID: 16759865 Review.
Cited by
-
Molecular regulation of antibiotic biosynthesis in streptomyces.Microbiol Mol Biol Rev. 2013 Mar;77(1):112-43. doi: 10.1128/MMBR.00054-12. Microbiol Mol Biol Rev. 2013. PMID: 23471619 Free PMC article. Review.
-
DNA mapping and kinetic modeling of the HrdB regulon in Streptomyces coelicolor.Nucleic Acids Res. 2019 Jan 25;47(2):621-633. doi: 10.1093/nar/gky1018. Nucleic Acids Res. 2019. PMID: 30371884 Free PMC article.
-
Metabolic and evolutionary insights into the closely-related species Streptomyces coelicolor and Streptomyces lividans deduced from high-resolution comparative genomic hybridization.BMC Genomics. 2010 Dec 1;11:682. doi: 10.1186/1471-2164-11-682. BMC Genomics. 2010. PMID: 21122120 Free PMC article.
-
Evolutionary dynamics of rhomboid proteases in Streptomycetes.BMC Res Notes. 2015 Jun 9;8:234. doi: 10.1186/s13104-015-1205-x. BMC Res Notes. 2015. PMID: 26054641 Free PMC article.
-
The Streptomyces coelicolor Small ORF trpM Stimulates Growth and Morphological Development and Exerts Opposite Effects on Actinorhodin and Calcium-Dependent Antibiotic Production.Front Microbiol. 2020 Feb 19;11:224. doi: 10.3389/fmicb.2020.00224. eCollection 2020. Front Microbiol. 2020. PMID: 32140146 Free PMC article.
References
-
- Abel, C. B. L., J. C. Lindon, D. Noble, B. A. M. Rudd, P. J. Sidebottom, and J. K. Nicholson. 1999. Characterization of metabolites in intact Streptomyces citricolor culture supernatants using high-resolution nuclear magnetic resonance and directly coupled high-pressure liquid chromatography-nuclear magnetic resonance spectroscopy. Anal. Biochem. 270:220-230. - PubMed
-
- Bentley, S. D., K. F. Chater, A.-M. Cerdeño-Tárraga, G. L. Challis, N. R. Thomson, K. D. James, D. E. Harris, M. A. Quail, H. Keiser, D. Harper, A. Bateman, S. Brown, G. Chandra, C. W. Chen, M. Collins, A. Cronin, A. Fraser, A. Goble, J. Hidalgo, T. Hornsby, S. Howarth, C.-H. Huang, T. Keiser, L. Larke, L. Murphy, K. Oliver, S. O'Neil, E. Rabbinowitsch, M.-A. Rajandream, K. Rutherford, S. Rutter, K. Seeger, D. Saunders, S. Sharp, R. Squares, S. Squares, K. Taylor, T. Warren, A. Wietzorrek, J. Woodward, B. G. Barrell, J. Parkhill, and D. A. Hopwood. 2002. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417:141-147. - PubMed
-
- Champness, W. 2000. Actinomycete development, antibiotic production, and phylogeny: questions and challenges, p. 11-31. In Y. V. Brun and L. J. Shimkets (ed.), Prokaryotic development. American Society for Microbiology, Washington, D.C.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

