Recognition of ferric catecholates by FepA
- PMID: 15150246
- PMCID: PMC415739
- DOI: 10.1128/JB.186.11.3578-3589.2004
Recognition of ferric catecholates by FepA
Abstract
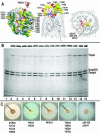
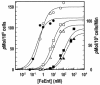
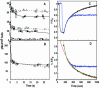
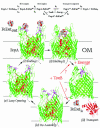
Escherichia coli FepA transports certain catecholate ferric siderophores, but not others, nor any noncatecholate compounds. Direct binding and competition experiments demonstrated that this selectivity originates during the adsorption stage. The synthetic tricatecholate Fe-TRENCAM bound to FepA with 50- to 100-fold-lower affinity than Fe-enterobactin (FeEnt), despite an identical metal center, and Fe-corynebactin only bound at much higher concentrations. Neither Fe-agrobactin nor ferrichrome bound at all, even at concentrations 10(6)-fold above the Kd. Thus, FepA only adsorbs catecholate iron complexes, and it selects FeEnt among even its close homologs. We used alanine scanning mutagenesis to study the contributions of surface aromatic residues to FeEnt recognition. Although not apparent from crystallography, aromatic residues in L3, L5, L7, L8, and L10 affected FepA's interaction with FeEnt. Among 10 substitutions that eliminated aromatic residues, Kd increased as much as 20-fold (Y481A and Y638A) and Km increased as much as 400-fold (Y478), showing the importance of aromaticity around the pore entrance. Although many mutations equally reduced binding and transport, others caused greater deficiencies in the latter. Y638A and Y478A increased Km 10- and 200-fold more, respectively, than Kd. N-domain loop deletions created the same phenotype: Delta60-67 (in NL1) and Delta98-105 (in NL2) increased Kd 10- to 20-fold but raised Km 500- to 700-fold. W101A (in NL2) had little effect on Kd but increased Km 1,000-fold. These data suggested that the primary role of the N terminus is in ligand uptake. Fluorescence and radioisotopic experiments showed biphasic release of FeEnt from FepA. In spectroscopic determinations, k(off1) was 0.03/s and k(off2) was 0.003/s. However, FepAY272AF329A did not manifest the rapid dissociation phase, corroborating the role of aromatic residues in the initial binding of FeEnt. Thus, the beta-barrel loops contain the principal ligand recognition determinants, and the N-domain loops perform a role in ligand transport.
Figures


Similar articles
-
Concerted loop motion triggers induced fit of FepA to ferric enterobactin.J Gen Physiol. 2014 Jul;144(1):71-80. doi: 10.1085/jgp.201311159. J Gen Physiol. 2014. PMID: 24981231 Free PMC article.
-
Conformational rearrangements in the N-domain of Escherichia coli FepA during ferric enterobactin transport.J Biol Chem. 2020 Apr 10;295(15):4974-4984. doi: 10.1074/jbc.RA119.011850. Epub 2020 Feb 25. J Biol Chem. 2020. PMID: 32098871 Free PMC article.
-
Aromatic components of two ferric enterobactin binding sites in Escherichia coli FepA.Mol Microbiol. 2000 Sep;37(6):1306-17. doi: 10.1046/j.1365-2958.2000.02093.x. Mol Microbiol. 2000. PMID: 10998164
-
Molecular mechanism of ferricsiderophore passage through the outer membrane receptor proteins of Escherichia coli.Biometals. 2007 Jun;20(3-4):263-74. doi: 10.1007/s10534-006-9060-9. Epub 2006 Dec 22. Biometals. 2007. PMID: 17186377 Review.
-
Three paradoxes of ferric enterobactin uptake.Front Biosci. 2003 Sep 1;8:s1422-36. doi: 10.2741/1233. Front Biosci. 2003. PMID: 12957833 Review.
Cited by
-
Siderophore-mediated iron acquisition by Klebsiella pneumoniae.J Bacteriol. 2024 May 23;206(5):e0002424. doi: 10.1128/jb.00024-24. Epub 2024 Apr 9. J Bacteriol. 2024. PMID: 38591913 Free PMC article.
-
FepA- and TonB-dependent bacteriophage H8: receptor binding and genomic sequence.J Bacteriol. 2007 Aug;189(15):5658-74. doi: 10.1128/JB.00437-07. Epub 2007 May 25. J Bacteriol. 2007. PMID: 17526714 Free PMC article.
-
Re-evaluation of the C-Glucosyltransferase IroB Illuminates Its Ability to C-Glucosylate Non-native Triscatecholate Enterobactin Mimics.Biochemistry. 2025 Jan 7;64(1):224-237. doi: 10.1021/acs.biochem.4c00581. Epub 2024 Dec 24. Biochemistry. 2025. PMID: 39718537
-
TonB-Dependent Receptor Repertoire of Pseudomonas aeruginosa for Uptake of Siderophore-Drug Conjugates.Antimicrob Agents Chemother. 2018 May 25;62(6):e00097-18. doi: 10.1128/AAC.00097-18. Print 2018 Jun. Antimicrob Agents Chemother. 2018. PMID: 29555629 Free PMC article.
-
Structure and Function of the PiuA and PirA Siderophore-Drug Receptors from Pseudomonas aeruginosa and Acinetobacter baumannii.Antimicrob Agents Chemother. 2017 Mar 24;61(4):e02531-16. doi: 10.1128/AAC.02531-16. Print 2017 Apr. Antimicrob Agents Chemother. 2017. PMID: 28137795 Free PMC article.
References
-
- Ames, G. F. 1974. Resolution of bacterial proteins by polyacrylamide gel electrophoresis on slabs: membrane, soluble, and periplasmic fractions. J. Biol. Chem. 249:634-644. - PubMed
-
- Armstrong, S. K., C. L. Francis, and M. A. McIntosh. 1990. Molecular analysis of the Escherichia coli ferric enterobactin receptor FepA. J. Biol. Chem. 265:14536-14543. - PubMed
-
- Braun, M., H. Killmann, and V. Braun. 1999. The β-barrel domain of FhuAΔ5-160 is sufficient for TonB-dependent FhuA activities of Escherichia coli. Mol. Microbiol. 33:1037-1049. - PubMed
-
- Braun, V. 1997. Surface signaling: novel transcription initiation mechanism starting from the cell surface. Arch. Microbiol. 167:325-331. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

