Simulation, experiment, and evolution: understanding nucleation in protein S6 folding
- PMID: 15150413
- PMCID: PMC420398
- DOI: 10.1073/pnas.0401672101
Simulation, experiment, and evolution: understanding nucleation in protein S6 folding
Abstract
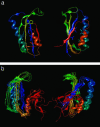
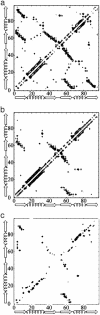
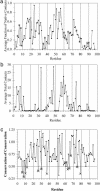
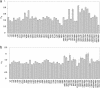
In this study, we explore nucleation and the transition state ensemble of the ribosomal protein S6 using a Monte Carlo (MC) Go model in conjunction with restraints from experiment. The results are analyzed in the context of extensive experimental and evolutionary data. The roles of individual residues in the folding nucleus are identified, and the order of events in the S6 folding mechanism is explored in detail. Interpretation of our results agrees with, and extends the utility of, experiments that shift phi-values by modulating denaturant concentration and presents strong evidence for the realism of the mechanistic details in our MC Go model and the structural interpretation of experimental phi-values. We also observe plasticity in the contacts of the hydrophobic core that support the specific nucleus. For S6, which binds to RNA and protein after folding, this plasticity may result from the conformational flexibility required to achieve biological function. These results present a theoretical and conceptual picture that is relevant in understanding the mechanism of nucleation in protein folding.
Figures
Similar articles
-
Folding of S6 structures with divergent amino acid composition: pathway flexibility within partly overlapping foldons.J Mol Biol. 2007 Jan 5;365(1):237-48. doi: 10.1016/j.jmb.2006.09.016. Epub 2006 Sep 12. J Mol Biol. 2007. PMID: 17056063
-
Common motifs and topological effects in the protein folding transition state.J Mol Biol. 2006 Jun 16;359(4):1075-85. doi: 10.1016/j.jmb.2006.04.015. Epub 2006 Apr 24. J Mol Biol. 2006. PMID: 16678203
-
Stabilization of the ribosomal protein S6 by trehalose is counterbalanced by the formation of a putative off-pathway species.J Mol Biol. 2005 Aug 12;351(2):402-16. doi: 10.1016/j.jmb.2005.05.056. J Mol Biol. 2005. PMID: 16002092
-
Protein folding theory: from lattice to all-atom models.Annu Rev Biophys Biomol Struct. 2001;30:361-96. doi: 10.1146/annurev.biophys.30.1.361. Annu Rev Biophys Biomol Struct. 2001. PMID: 11340064 Review.
-
Protein folding simulations: from coarse-grained model to all-atom model.IUBMB Life. 2009 Jun;61(6):627-43. doi: 10.1002/iub.223. IUBMB Life. 2009. PMID: 19472192 Review.
Cited by
-
Folding of Cu/Zn superoxide dismutase suggests structural hotspots for gain of neurotoxic function in ALS: parallels to precursors in amyloid disease.Proc Natl Acad Sci U S A. 2006 Jul 5;103(27):10218-10223. doi: 10.1073/pnas.0601696103. Epub 2006 Jun 23. Proc Natl Acad Sci U S A. 2006. PMID: 16798882 Free PMC article.
-
Balancing energy and entropy: a minimalist model for the characterization of protein folding landscapes.Proc Natl Acad Sci U S A. 2005 Jul 19;102(29):10141-6. doi: 10.1073/pnas.0409471102. Epub 2005 Jul 8. Proc Natl Acad Sci U S A. 2005. PMID: 16006532 Free PMC article.
-
Coevolution of function and the folding landscape: correlation with density of native contacts.Biophys J. 2008 Nov 1;95(9):L57-9. doi: 10.1529/biophysj.108.143388. Epub 2008 Aug 15. Biophys J. 2008. PMID: 18708465 Free PMC article.
-
Analysis of core-periphery organization in protein contact networks reveals groups of structurally and functionally critical residues.J Biosci. 2015 Oct;40(4):683-99. doi: 10.1007/s12038-015-9554-0. J Biosci. 2015. PMID: 26564971
-
Insights from coarse-grained Gō models for protein folding and dynamics.Int J Mol Sci. 2009 Mar;10(3):889-905. doi: 10.3390/ijms10030889. Epub 2009 Mar 2. Int J Mol Sci. 2009. PMID: 19399227 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

