Structural basis for nicotinamide cleavage and ADP-ribose transfer by NAD(+)-dependent Sir2 histone/protein deacetylases
- PMID: 15150415
- PMCID: PMC423234
- DOI: 10.1073/pnas.0401057101
Structural basis for nicotinamide cleavage and ADP-ribose transfer by NAD(+)-dependent Sir2 histone/protein deacetylases
Abstract
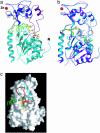
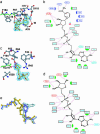
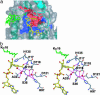
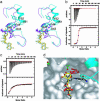
Sir2 enzymes are broadly conserved from bacteria to humans and have been implicated to play roles in gene silencing, DNA repair, genome stability, longevity, metabolism, and cell physiology. These enzymes bind NAD(+) and acetyllysine within protein targets and generate lysine, 2'-O-acetyl-ADP-ribose, and nicotinamide products. To provide structural insights into the chemistry catalyzed by Sir2 proteins we report the high-resolution ternary structure of yeast Hst2 (homologue of Sir two 2) with an acetyllysine histone H4 peptide and a nonhydrolyzable NAD(+) analogue, carba-NAD(+), as well as an analogous ternary complex with a reaction intermediate analog formed immediately after nicotinamide hydrolysis, ADP-ribose. The ternary complex with carba-NAD(+) reveals that the nicotinamide group makes stabilizing interactions within a binding pocket harboring conserved Sir2 residues. Moreover, an asparagine residue, N116, strictly conserved within Sir2 proteins and shown to be essential for nicotinamide exchange, is in position to stabilize the oxocarbenium intermediate that has been proposed to proceed the hydrolysis of nicotinamide. A comparison of this structure with the ADP-ribose ternary complex and a previously reported ternary complex with the 2'-O-acetyl-ADP-ribose reaction product reveals that the ribose ring of the cofactor and the highly conserved beta1-alpha2 loop of the protein undergo significant structural rearrangements to facilitate the ordered NAD(+) reactions of nicotinamide cleavage and ADP-ribose transfer to acetate. Together, these studies provide insights into the chemistry of NAD(+) cleavage and acetylation by Sir2 proteins and have implications for the design of Sir2-specific regulatory molecules.
Figures

Similar articles
-
Substrate specificity and kinetic mechanism of the Sir2 family of NAD+-dependent histone/protein deacetylases.Biochemistry. 2004 Aug 3;43(30):9877-87. doi: 10.1021/bi049592e. Biochemistry. 2004. PMID: 15274642
-
Sir2 regulation by nicotinamide results from switching between base exchange and deacetylation chemistry.Biochemistry. 2003 Aug 12;42(31):9249-56. doi: 10.1021/bi034959l. Biochemistry. 2003. PMID: 12899610
-
SIR2: the biochemical mechanism of NAD(+)-dependent protein deacetylation and ADP-ribosyl enzyme intermediates.Curr Med Chem. 2004 Apr;11(7):807-26. doi: 10.2174/0929867043455675. Curr Med Chem. 2004. PMID: 15078167 Review.
-
Role of NAD(+) in the deacetylase activity of the SIR2-like proteins.Biochem Biophys Res Commun. 2000 Nov 30;278(3):685-90. doi: 10.1006/bbrc.2000.3854. Biochem Biophys Res Commun. 2000. PMID: 11095969
-
Structure and chemistry of the Sir2 family of NAD+-dependent histone/protein deactylases.Biochem Soc Trans. 2004 Dec;32(Pt 6):904-9. doi: 10.1042/BST0320904. Biochem Soc Trans. 2004. PMID: 15506920 Review.
Cited by
-
Bypassing the catalytic activity of SIR2 for SIR protein spreading in Saccharomyces cerevisiae.Mol Biol Cell. 2006 Dec;17(12):5287-97. doi: 10.1091/mbc.e06-08-0669. Epub 2006 Oct 11. Mol Biol Cell. 2006. PMID: 17035629 Free PMC article.
-
Nuclear ADP-ribosylation reactions in mammalian cells: where are we today and where are we going?Microbiol Mol Biol Rev. 2006 Sep;70(3):789-829. doi: 10.1128/MMBR.00040-05. Microbiol Mol Biol Rev. 2006. PMID: 16959969 Free PMC article. Review.
-
Structure and biochemical functions of SIRT6.J Biol Chem. 2011 Apr 22;286(16):14575-87. doi: 10.1074/jbc.M111.218990. Epub 2011 Mar 1. J Biol Chem. 2011. PMID: 21362626 Free PMC article.
-
Characterization of CobB kinetics and inhibition by nicotinamide.PLoS One. 2017 Dec 18;12(12):e0189689. doi: 10.1371/journal.pone.0189689. eCollection 2017. PLoS One. 2017. PMID: 29253849 Free PMC article.
-
Nampt/PBEF/Visfatin: a regulator of mammalian health and longevity?Exp Gerontol. 2006 Aug;41(8):718-26. doi: 10.1016/j.exger.2006.06.003. Epub 2006 Jul 13. Exp Gerontol. 2006. PMID: 16842957 Free PMC article. Review.
References
-
- Guarente, L. (2000) Genes Dev. 14, 1021–1026. - PubMed
-
- Avalos, J. L., Celic, I., Muhammad, S., Cosgrove, M. S., Boeke, J. D. & Wolberger, C. (2002) Mol. Cell 10, 523–535. - PubMed
-
- Finnin, M. S., Donigian, J. R. & Pavletich, N. P. (2001) Nat. Struct. Biol. 8, 621–625. - PubMed
-
- Zhao, K., Chai, X., Clements, A. & Marmorstein, R. (2003) Nat. Struct. Biol. 10, 864–871. - PubMed
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

