Severe acute respiratory syndrome coronavirus (SARS-CoV) infection inhibition using spike protein heptad repeat-derived peptides
- PMID: 15150417
- PMCID: PMC420415
- DOI: 10.1073/pnas.0400576101
Severe acute respiratory syndrome coronavirus (SARS-CoV) infection inhibition using spike protein heptad repeat-derived peptides
Abstract
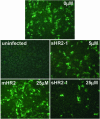
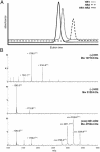
The coronavirus SARS-CoV is the primary cause of the life-threatening severe acute respiratory syndrome (SARS). With the aim of developing therapeutic agents, we have tested peptides derived from the membrane-proximal (HR2) and membrane-distal (HR1) heptad repeat region of the spike protein as inhibitors of SARS-CoV infection of Vero cells. It appeared that HR2 peptides, but not HR1 peptides, were inhibitory. Their efficacy was, however, significantly lower than that of corresponding HR2 peptides of the murine coronavirus mouse hepatitis virus (MHV) in inhibiting MHV infection. Biochemical and electron microscopical analyses showed that, when mixed, SARS-CoV HR1 and HR2 peptides assemble into a six-helix bundle consisting of HR1 as a central triple-stranded coiled coil in association with three HR2 alpha-helices oriented in an antiparallel manner. The stability of this complex, as measured by its resistance to heat dissociation, appeared to be much lower than that of the corresponding MHV complex, which may explain the different inhibitory potencies of the HR2 peptides. Analogous to other class I viral fusion proteins, the six-helix complex supposedly represents a postfusion conformation that is formed after insertion of the fusion peptide, proposed here for coronaviruses to be located immediately upstream of HR1, into the target membrane. The resulting close apposition of fusion peptide and spike transmembrane domain facilitates membrane fusion. The inhibitory potency of the SARS-CoV HR2-peptides provides an attractive basis for the development of a therapeutic drug for SARS.
Figures



References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

