Alteration of the copy number and deletion of mitochondrial DNA in human hepatocellular carcinoma
- PMID: 15150555
- PMCID: PMC2409531
- DOI: 10.1038/sj.bjc.6601838
Alteration of the copy number and deletion of mitochondrial DNA in human hepatocellular carcinoma
Abstract
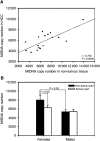
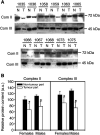
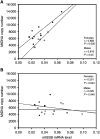
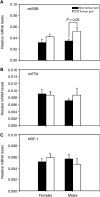
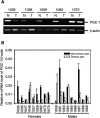
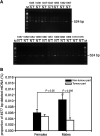
Somatic mutations in mitochondrial DNA (mtDNA) have been detected in hepatocellular carcinoma (HCC). However, it remains unclear whether mtDNA copy number and mitochondrial biogenesis are altered in HCC. In this study, we found that mtDNA copy number and the content of mitochondrial respiratory proteins were reduced in HCCs as compared with the corresponding non-tumorous livers. MtDNA copy number was significantly reduced in female HCC but not in male HCC. Expression of the peroxisome proliferator-activated receptor gamma coactivator-1 was significantly repressed in HCCs (P<0.005), while the expression of the mitochondrial single-strand DNA-binding protein was upregulated, indicating that the regulation of mitochondria biogenesis is disturbed in HCC. Moreover, 22% of HCCs carried a somatic mutation in the mtDNA D-loop region. The non-tumorous liver of the HCC patients with a long-term alcohol-drinking history contained reduced mtDNA copy number (P<0.05) and higher level of the 4977 bp-deleted mtDNA (P<0.05) as compared with non-alcohol patients. Our results suggest that reduced mtDNA copy number, impaired mitochondrial biogenesis and somatic mutations in mtDNA are important events during carcinogenesis of HCC, and the differential alterations in mtDNA of male and female HCC may contribute to the differences in the clinical manifestation between female and male HCC patients.
Figures

References
-
- Anderson S, Bankier AT, Barrell BG, De Bruijn MHL, Coulson AR, Drouin J, Eperon IC, Nierlich DP, Roe BA, Sanger F, Schreier PH, Smith AJH, Staden R, Young IG (1981) Sequence and organization of the human mitochondrial genome. Nature 290: 457–465 - PubMed
-
- Augenlicht LH, Heerdt BG (2001) Mitochondria: integrators in tumorigenesis? Nat Genet 28: 104–105 - PubMed
-
- Cederbaum AI (1999) Effects of alcohol on hepatic mitochondrial function and DNA. Gastroenterology 117: 265–269 - PubMed
-
- Chen CJ, Yu MW, Liaw YF (1997) Epidemiological characteristics and risk factors of hepatocellular carcinoma. J Gastroenterol Hepatol 12: S294–S308 - PubMed
-
- Cuezva JM, Krajewska M, De Heredia ML, Krajewski S, Santamaria G, Kim H, Zapata JM (2002) The bioenergetic signature of cancer: a marker of tumor progression. Cancer Res 62: 6674–6681 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

