The ankyrin repeat as molecular architecture for protein recognition
- PMID: 15152081
- PMCID: PMC2279977
- DOI: 10.1110/ps.03554604
The ankyrin repeat as molecular architecture for protein recognition
Abstract
The ankyrin repeat is one of the most frequently observed amino acid motifs in protein databases. This protein-protein interaction module is involved in a diverse set of cellular functions, and consequently, defects in ankyrin repeat proteins have been found in a number of human diseases. Recent biophysical, crystallographic, and NMR studies have been used to measure the stability and define the various topological features of this motif in an effort to understand the structural basis of ankyrin repeat-mediated protein-protein interactions. Characterization of the folding and assembly pathways suggests that ankyrin repeat domains generally undergo a two-state folding transition despite their modular structure. Also, the large number of available sequences has allowed the ankyrin repeat to be used as a template for consensus-based protein design. Such projects have been successful in revealing positions responsible for structure and function in the ankyrin repeat as well as creating a potential universal scaffold for molecular recognition.
Figures
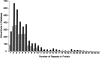

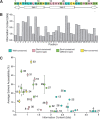
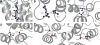
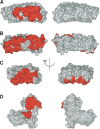
References
-
- Bae, J., Donigian, J.R., and Hsueh, A.J.W. 2003. Tankyrase 1 interacts with Mcl-1 proteins and inhibits their regulation of apoptosis. J. Biol. Chem. 278 5195–5204. - PubMed
-
- Batchelor, A.H., Piper, D.E., de la Brousse, F.C., McKnight, S.L., and Wolberger, C. 1998. The structure of GABPα/β: An ETS domain-ankyrin repeat heterodimer bound to DNA. Science 279 1037–1041. - PubMed
-
- Becker, T.M., Rizos, H., Kefford, R.F., and Mann, G.J. 2001. Functional impairment of melanoma-associated p16INK4a mutants in melanoma cells despite retention of cyclin-dependent kinase 4 binding. Clin. Cancer Res. 7 3282–3288. - PubMed
-
- Binz, H.K., Stumpp, M.T., Forrer, P., Amstutz, P., and Pluckthun, A. 2003. Designing repeat proteins: Well-expressed, soluble and stable proteins from combinatorial libraries of consensus ankyrin repeat proteins. J. Mol. Biol. 332 489–503. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

