A second human antiretroviral factor, APOBEC3F, is suppressed by the HIV-1 and HIV-2 Vif proteins
- PMID: 15152192
- PMCID: PMC423288
- DOI: 10.1038/sj.emboj.7600246
A second human antiretroviral factor, APOBEC3F, is suppressed by the HIV-1 and HIV-2 Vif proteins
Abstract
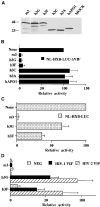
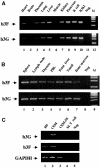
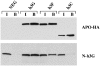
The HIV-1 Vif protein suppresses the inhibition of viral replication caused by the human antiretroviral factor APOBEC3G. As a result, HIV-1 mutants that do not express the Vif protein are replication incompetent in 'nonpermissive' cells, such as primary T cells and the T-cell line CEM, that express APOBEC3G. In contrast, Vif-defective HIV-1 replicates effectively in 'permissive' cell lines, such as a derivative of CEM termed CEM-SS, that do not express APOBEC3G. Here, we show that a second human protein, APOBEC3F, is also specifically packaged into HIV-1 virions and inhibits their infectivity. APOBEC3F binds the HIV-1 Vif protein specifically and Vif suppresses both the inhibition of virus infectivity caused by APOBEC3F and virion incorporation of APOBEC3F. Surprisingly, APOBEC3F and APOBEC3G are extensively coexpressed in nonpermissive human cells, including primary lymphocytes and the cell line CEM, where they form heterodimers. In contrast, both genes are quiescent in the permissive CEM derivative CEM-SS. Together, these data argue that HIV-1 Vif has evolved to suppress at least two distinct but related human antiretroviral DNA-editing enzymes.
Figures


References
-
- Beale RCL, Petersen-Mahrt SK, Watt IN, Harris RS, Rada C, Neuberger MS (2004) Comparison of the differential context-dependence of DNA deamination by APOBEC enzymes: correlation with mutation spectra in vivo. J Mol Biol 337: 585–596 - PubMed
-
- Borman AM, Quillent C, Charneau P, Kean KM, Clavel F (1995) A highly defective HIV-1 group O provirus: evidence for the role of local sequence determinants in G → A hypermutation during negative-strand viral DNA synthesis. Virology 208: 601–609 - PubMed
-
- Chesebro B, Wehrly K, Nishio J, Perryman S (1992) Macrophage-tropic human immunodeficiency virus isolates from different patients exhibit unusual V3 envelope sequence homogeneity in comparison with T-cell-tropic isolates: definition of critical amino acids involved in cell tropism. J Virol 66: 6547–6554 - PMC - PubMed
-
- Conticello SG, Harris RS, Neuberger MS (2003) The Vif protein of HIV triggers degradation of the human antiretroviral DNA deaminase APOBEC3G. Curr Biol 13: 2009–2013 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

