Nosocomial spread of ceftazidime-resistant Klebsiella pneumoniae strains producing a novel class a beta-lactamase, GES-3, in a neonatal intensive care unit in Japan
- PMID: 15155185
- PMCID: PMC415581
- DOI: 10.1128/AAC.48.6.1960-1967.2004
Nosocomial spread of ceftazidime-resistant Klebsiella pneumoniae strains producing a novel class a beta-lactamase, GES-3, in a neonatal intensive care unit in Japan
Abstract
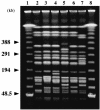
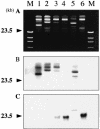
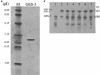
Klebsiella pneumoniae strain KG525, which showed high-level resistance to broad-spectrum cephalosporins, was isolated from the neonatal intensive care unit (NICU) of a Japanese hospital in March 2002. The ceftazidime resistance of strain KG525 was transferable to Escherichia coli CSH-2 by conjugation. Cloning and sequence analysis revealed that production of a novel extended-spectrum class A beta-lactamase (pI 7.0), designated GES-3, which had two amino acid substitutions of M62T and E104K on the basis of the sequence of GES-1, was responsible for resistance in strain KG525 and its transconjugant. The bla(GES-3) gene was located as the first gene cassette in a class 1 integron that also contained an aacA1-orfG fused gene cassette and one unique cassette that has not been described in other class 1 integrons and ended with a truncated 3' conserved segment by insertion of IS26. Another five ceftazidime-resistant K. pneumoniae strains, strains KG914, KG1116, KG545, KG502, and KG827, which were isolated from different neonates during a 1-year period in the same NICU where strain KG525 had been isolated, were also positive for GES-type beta-lactamase genes by PCR. Pulsed-field gel electrophoresis and enterobacterial repetitive intergenic consensus-PCR analyses displayed genetic relatedness among the six K. pneumoniae strains. Southern hybridization analysis with a GES-type beta-lactamase gene-specific probe showed that the locations of bla(GES) were multiple and diverse among the six strains. These findings suggest that within the NICU setting genetically related K. pneumoniae strains carrying the bla(GES) gene were ambushed with genetic rearrangements that caused the multiplication and translocation of the bla(GES) gene.
Figures

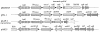
Comment in
-
Nomenclature of GES-type extended-spectrum beta-lactamases.Antimicrob Agents Chemother. 2005 May;49(5):2148; author reply 2148-50. doi: 10.1128/AAC.49.5.2148-2150.2005. Antimicrob Agents Chemother. 2005. PMID: 15855553 Free PMC article. No abstract available.
Similar articles
-
Molecular characterization of a cephamycin-hydrolyzing and inhibitor-resistant class A beta-lactamase, GES-4, possessing a single G170S substitution in the omega-loop.Antimicrob Agents Chemother. 2004 Aug;48(8):2905-10. doi: 10.1128/AAC.48.8.2905-2910.2004. Antimicrob Agents Chemother. 2004. PMID: 15273099 Free PMC article.
-
Emergence of Klebsiella pneumoniae carrying the novel extended-spectrum beta-lactamase gene variants bla(SHV-40), bla(TEM-116) and the class 1 integron-associated bla(GES-7) in Brazil.Clin Microbiol Infect. 2010 Jun;16(6):630-2. doi: 10.1111/j.1469-0691.2009.02944.x. Epub 2009 Aug 18. Clin Microbiol Infect. 2010. PMID: 19689462
-
Risk factors for extended-spectrum beta-lactamase-producing Serratia marcescens and Klebsiella pneumoniae acquisition in a neonatal intensive care unit.J Hosp Infect. 2007 Oct;67(2):135-41. doi: 10.1016/j.jhin.2007.07.026. Epub 2007 Sep 19. J Hosp Infect. 2007. PMID: 17884248
-
High rates of metallo-beta-lactamase-producing Klebsiella pneumoniae in Greece--a review of the current evidence.Euro Surveill. 2008 Jan 24;13(4):8023. Euro Surveill. 2008. PMID: 18445397 Review.
-
Klebsiella pneumoniae and other Enterobacteriaceae producing novel plasmid-mediated beta-lactamases markedly active against third-generation cephalosporins: epidemiologic studies.Rev Infect Dis. 1988 Jul-Aug;10(4):850-9. doi: 10.1093/clinids/10.4.850. Rev Infect Dis. 1988. PMID: 3055177 Review.
Cited by
-
Drug resistance phenotypes and genotypes in Mexico in representative gram-negative species: Results from the infivar network.PLoS One. 2021 Mar 17;16(3):e0248614. doi: 10.1371/journal.pone.0248614. eCollection 2021. PLoS One. 2021. PMID: 33730101 Free PMC article.
-
Rapid detection and sequence-specific differentiation of extended-spectrum beta-lactamase GES-2 from Pseudomonas aeruginosa by use of a real-time PCR assay.Antimicrob Agents Chemother. 2004 Oct;48(10):4059-62. doi: 10.1128/AAC.48.10.4059-4062.2004. Antimicrob Agents Chemother. 2004. PMID: 15388481 Free PMC article.
-
Complete genome sequence, lifestyle, and multi-drug resistance of the human pathogen Corynebacterium resistens DSM 45100 isolated from blood samples of a leukemia patient.BMC Genomics. 2012 Apr 23;13:141. doi: 10.1186/1471-2164-13-141. BMC Genomics. 2012. PMID: 22524407 Free PMC article.
-
Pyrosequencing as a rapid tool for identification of GES-type extended-spectrum beta-lactamases.J Clin Microbiol. 2006 Aug;44(8):3008-11. doi: 10.1128/JCM.02576-05. J Clin Microbiol. 2006. PMID: 16891529 Free PMC article.
-
Detection of Carbapenemase Production in Gram-negative Bacteria.J Lab Physicians. 2014 Jul;6(2):69-75. doi: 10.4103/0974-2727.141497. J Lab Physicians. 2014. PMID: 25328329 Free PMC article.
References
-
- Arakawa, Y., M. Ohta, N. Kido, Y. Fujii, T. Komatsu, and N. Kato. 1986. Close evolutionary relationship between the chromosomally encoded βlactamase gene of Klebsiella pneumoniae and the TEM β-lactamase gene mediated by R-plasmids. FEBS Lett. 207:69-74. - PubMed
-
- Bradford, P. A., C. E. Cherubin, V. Idemyor, B. A. Rasmussen, and K. Bush. 1994. Multiply resistant Klebsiella pneumoniae strains from two Chicago hospitals: identification of the extended-spectrum TEM-12 and TEM-10 ceftazidime-hydrolyzing β-lactamases in a single isolate. Antimicrob. Agents Chemother. 38:761-766. - PMC - PubMed
-
- Bradford, P. A., C. Urban, N. Mariano, S. J. Projan, J. J. Rahal, and K. Bush. 1997. Imipenem resistance in Klebsiella pneumoniae is associated with the combination of ACT-1, a plasmid-mediated AmpC β-lactamase, and the loss of an outer membrane protein. Antimicrob. Agents Chemother. 41:563-569. - PMC - PubMed
-
- Burwen, D. R., S. N. Banerjee, R. P. Gaynes, et al. 1994. Ceftazidime resistance among selected nosocomial gram-negative bacilli in the United States. J. Infect. Dis. 170:1622-1625. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

