Ciprofloxacin dimers target gyrase in Streptococcus pneumoniae
- PMID: 15155208
- PMCID: PMC415600
- DOI: 10.1128/AAC.48.6.2108-2115.2004
Ciprofloxacin dimers target gyrase in Streptococcus pneumoniae
Abstract
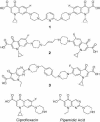
We have examined the antipneumococcal activities of novel quinolone dimers in which ciprofloxacin was tethered to itself or to pipemidic acid by linkage of C-7 piperazinyl rings. Symmetric 2,6-lutidinyl- and trans-butenyl-linked ciprofloxacin dimers (dimers 1 and 2, respectively) and a pipemidic acid-ciprofloxacin dimer (dimer 3) had activities against Streptococcus pneumoniae strain 7785 that were comparable to that of ciprofloxacin, i.e., MICs of 2, 1, and 4 to 8 microg/ml versus an MIC of 1 to 2 microg/ml, respectively. Surprisingly, unlike ciprofloxacin (which targets topoisomerase IV), several lines of evidence revealed that the dimers act through gyrase in S. pneumoniae. First, ciprofloxacin-resistant parC mutants of strain 7785 remained susceptible to dimers 1 to 3, whereas a gyrA mutation conferred a four- to eightfold increase in the dimer MIC but had little effect on ciprofloxacin activity. Second, dimer 1 selected first-step gyrA (S81Y or S81F) mutants (MICs, 8 to 16 microg/ml) that carried wild-type topoisomerase IV parE-parC genes. Third, dimers 1 and 2 promoted comparable DNA cleavage by S. pneumoniae gyrase and topoisomerase IV, whereas ciprofloxacin-mediated cleavage was 10-fold more efficient with topoisomerase IV than with gyrase. Fourth, the GyrA S81F and ParC S79F enzymes were resistant to dimers, confirming that the resistance phenotype is largely silent in parC mutants. Although a dimer molecule could bind very tightly by bridging quinolone binding sites in the enzyme-DNA complex, the greater potency of ciprofloxacin against gyrase and topoisomerase IV suggests that dimers 1 to 3 bind in a monomeric fashion. The bulky C-7 side chain may explain dimer targeting of gyrase and activity against efflux mutants. Tethered quinolones have potential as mechanistic tools and as novel antimicrobial agents.
Figures




Similar articles
-
Quinolone resistance mutations in Streptococcus pneumoniae GyrA and ParC proteins: mechanistic insights into quinolone action from enzymatic analysis, intracellular levels, and phenotypes of wild-type and mutant proteins.Antimicrob Agents Chemother. 2001 Nov;45(11):3140-7. doi: 10.1128/AAC.45.11.3140-3147.2001. Antimicrob Agents Chemother. 2001. PMID: 11600369 Free PMC article.
-
Cleavable-complex formation by wild-type and quinolone-resistant Streptococcus pneumoniae type II topoisomerases mediated by gemifloxacin and other fluoroquinolones.Antimicrob Agents Chemother. 2002 Feb;46(2):413-9. doi: 10.1128/AAC.46.2.413-419.2002. Antimicrob Agents Chemother. 2002. PMID: 11796351 Free PMC article.
-
Engineering the specificity of antibacterial fluoroquinolones: benzenesulfonamide modifications at C-7 of ciprofloxacin change its primary target in Streptococcus pneumoniae from topoisomerase IV to gyrase.Antimicrob Agents Chemother. 2000 Feb;44(2):320-5. doi: 10.1128/AAC.44.2.320-325.2000. Antimicrob Agents Chemother. 2000. PMID: 10639357 Free PMC article.
-
Quinolone mode of action.Drugs. 1995;49 Suppl 2:10-5. doi: 10.2165/00003495-199500492-00004. Drugs. 1995. PMID: 8549276 Review.
-
Dual activity of fluoroquinolones against Streptococcus pneumoniae: the facts behind the claims.J Antimicrob Chemother. 2002 Jun;49(6):893-5. doi: 10.1093/jac/dkf047. J Antimicrob Chemother. 2002. PMID: 12039880 Review. No abstract available.
Cited by
-
Synthesis and evaluation of 1-cyclopropyl-2-thioalkyl-8-methoxy fluoroquinolones.Bioorg Med Chem Lett. 2011 Aug 1;21(15):4585-8. doi: 10.1016/j.bmcl.2011.05.112. Epub 2011 Jun 6. Bioorg Med Chem Lett. 2011. PMID: 21705218 Free PMC article.
-
Efficient Synthesis of the 2-amino-6-chloro-4-cyclopropyl-7-fluoro-5-methoxy-pyrido[1,2-c]pyrimidine-1,3-dione core ring system.Tetrahedron Lett. 2009 Feb 18;50(7):785-789. doi: 10.1016/j.tetlet.2008.11.121. Tetrahedron Lett. 2009. PMID: 20160840 Free PMC article.
-
In vitro evaluation of CBR-2092, a novel rifamycin-quinolone hybrid antibiotic: studies of the mode of action in Staphylococcus aureus.Antimicrob Agents Chemother. 2008 Jul;52(7):2313-23. doi: 10.1128/AAC.01649-07. Epub 2008 Apr 28. Antimicrob Agents Chemother. 2008. PMID: 18443108 Free PMC article.
-
A Novel indole compound that inhibits Pseudomonas aeruginosa growth by targeting MreB is a substrate for MexAB-OprM.J Bacteriol. 2007 Oct;189(19):6870-81. doi: 10.1128/JB.00805-07. Epub 2007 Jul 20. J Bacteriol. 2007. PMID: 17644596 Free PMC article.
-
Fluoroquinolone-Transition Metal Complexes: A Strategy to Overcome Bacterial Resistance.Microorganisms. 2021 Jul 14;9(7):1506. doi: 10.3390/microorganisms9071506. Microorganisms. 2021. PMID: 34361943 Free PMC article. Review.
References
-
- Adams, D. E., E. M. Shekman, E. L. Zechiedrich, M. B. Schmid, and N. R. Cozzarelli. 1992. The role of topoisomerase IV in partitioning DNA replicons and the structure of catenated intermediates in DNA replication. Cell 71:277-288. - PubMed
-
- Alovero, F. L., X.-S. Pan, J. E. Morris, R. H. Manzo, and L. M. Fisher. 2000. Engineering the specificity of antibacterial fluoroquinolones: benzenesulfonamide modifications at C-7 of ciprofloxacin change its primary target in Streptococcus pneumoniae from topoisomerase IV to gyrase. Antimicrob. Agents Chemother. 44:320-325. - PMC - PubMed
-
- Ferrero, L., B. Cameron, B. Manse, D. Lagneux, J. Crouzet, A. Famechon, and F. Blanche. 1994. Cloning and primary structure of Staphylococcus aureus DNA topoisomerase IV: a primary target of fluoroquinolones. Mol. Microbiol. 13:641-653. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

