Arabidopsis downy mildew resistance gene RPP27 encodes a receptor-like protein similar to CLAVATA2 and tomato Cf-9
- PMID: 15155873
- PMCID: PMC514143
- DOI: 10.1104/pp.103.037770
Arabidopsis downy mildew resistance gene RPP27 encodes a receptor-like protein similar to CLAVATA2 and tomato Cf-9
Retraction in
-
Arabidopsis downy mildew resistance gene RPP27 encodes a receptor-like protein similar to CLAVATA2 and tomato Cf-9.Plant Physiol. 2007 Feb;143(2):1079. doi: 10.1104/pp.104.900215. Plant Physiol. 2007. PMID: 17284584 Free PMC article. No abstract available.
Abstract
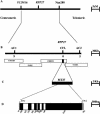
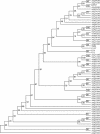
The Arabidopsis Ler-RPP27 gene confers AtSgt1b-independent resistance to downy mildew (Peronospora parasitica) isolate Hiks1. The RPP27 locus was mapped to a four-bacterial artificial chromosome interval on chromosome 1 from genetic analysis of a cross between the enhanced susceptibility mutant Col-edm1 (Col-sgt1) and Landsberg erecta (Ler-0). A Cf-like candidate gene in this interval was PCR amplified from Ler-0 and transformed into mutant Col-rpp7.1 plants. Homozygous transgenic lines conferred resistance to Hiks1 and at least four Ler-0 avirulent/Columbia-0 (Col-0) virulent isolates of downy mildew pathogen. A full-length RPP27 cDNA was isolated, and analysis of the deduced amino acid sequences showed that the gene encodes a receptor-like protein (RLP) with a distinct domain structure, composed of a signal peptide followed by extracellular Leu-rich repeats, a membrane spanning region, and a short cytoplasmic carboxyl domain. RPP27 is the first RLP-encoding gene to be implicated in disease resistance in Arabidopsis, enabling the deployment of Arabidopsis techniques to investigate the mechanisms of RLP function. Homology searches of the Arabidopsis genome, using the RPP27, Cf-9, and Cf-2 protein sequences as a starting point, identify 59 RLPs, including the already known CLAVATA2 and TOO MANY MOUTHS genes. A combination of sequence and phylogenetic analysis of these predicted RLPs reveals conserved structural features of the family.
Figures




References
-
- Adachi J, Hasegawa M (1996) Molphy Version 2.3: Programs for Molecular Phylogenetics Based on Maximum Likelihood. Computer Science Monographs, No. 28. Institute of Statistical Mathematics, Tokyo
-
- Bittner-Eddy PD, Crute IR, Holub EB, Beynon JL (2000) RPP13 is a simple locus in Arabidopsis thaliana for alleles that specify downy mildew resistance to different avirulence determinants in Peronospora parasitica. Plant J 21: 177–188 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

