NAI1 gene encodes a basic-helix-loop-helix-type putative transcription factor that regulates the formation of an endoplasmic reticulum-derived structure, the ER body
- PMID: 15155889
- PMCID: PMC490044
- DOI: 10.1105/tpc.021154
NAI1 gene encodes a basic-helix-loop-helix-type putative transcription factor that regulates the formation of an endoplasmic reticulum-derived structure, the ER body
Abstract
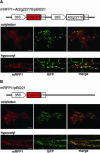
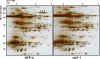
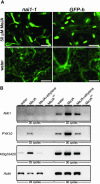
Plant cells develop various types of endoplasmic reticulum (ER)-derived structures with specific functions. ER body, an ER-derived compartment in Arabidopsis thaliana, is a spindle-shaped structure. The NAI1 gene regulates the development of ER bodies because mutation of NAI1 abolishes the formation of ER bodies. To better understand the role of NAI1, we cloned the NAI1 gene using a positional cloning strategy. The nai1-1 mutant had a single nucleotide change at an intron acceptor site of At2g22770 (NAI1 gene). Because of this mutation, aberrant splicing of NAI1 mRNA occurs in the nai1-1 mutant. NAI1 encodes a transcription factor that has a basic-helix-loop-helix (bHLH) domain. Transient expression of NAI1 induced ER bodies in the nai1-1 mutant. Two-dimensional electrophoresis and RT-PCR analyses showed that a putative lectin was depressed at both the mRNA and protein levels in nai1 mutants, as was a beta-glucosidase (PYK10). Our results provide direct evidence that a bHLH protein plays a role in the formation of ER bodies.
Copyright 2004 American Society of Plant Biologists
Figures






References
-
- Augur, C., Benhamou, N., Darvill, A., and Albersheim, P. (1993). Purification, characterization, and cell wall localization of an α-fucosidase that inactivates a xyloglucan oligosaccarin. Plant J. 3, 415–426. - PubMed
-
- Babcock, G.D., and Esen, A. (1994). Substrate specificity of maize β-glucosidase. Plant Sci. 101, 31–39.
-
- Baumann, O., and Walz, B. (2001). Endoplasmic reticulum of animal cells and its organization into structural and functional domains. Int. Rev. Cytol. 205, 149–214. - PubMed
-
- Behnke, H.-D., and Eschlbeck, G. (1978). Dilated cisternae in Capparales—An attempt towards the characterization of a specific endoplasmic reticulum. Protoplasma 97, 351–363.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

