Evidence for assembly of prions with left-handed beta-helices into trimers
- PMID: 15155909
- PMCID: PMC420396
- DOI: 10.1073/pnas.0402254101
Evidence for assembly of prions with left-handed beta-helices into trimers
Abstract
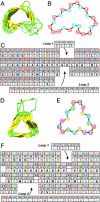
Studies using low-resolution fiber diffraction, electron microscopy, and atomic force microscopy on various amyloid fibrils indicate that the misfolded conformers must be modular, compact, and adopt a cross-beta structure. In an earlier study, we used electron crystallography to delineate molecular models of the N-terminally truncated, disease-causing isoform (PrP(Sc)) of the prion protein, designated PrP 27-30, which polymerizes into amyloid fibrils, but we were unable to choose between a trimeric or hexameric arrangement of right- or left-handed beta-helical models. From a study of 119 all-beta folds observed in globular proteins, we have now determined that, if PrP(Sc) follows a known protein fold, it adopts either a beta-sandwich or parallel beta-helical architecture. With increasing evidence arguing for a parallel beta-sheet organization in amyloids, we contend that the sequence of PrP is compatible with a parallel left-handed beta-helical fold. Left-handed beta-helices readily form trimers, providing a natural template for a trimeric model of PrP(Sc). This trimeric model accommodates the PrP sequence from residues 89-175 in a beta-helical conformation with the C terminus (residues 176-227), retaining the disulfide-linked alpha-helical conformation observed in the normal cellular isoform. In addition, the proposed model matches the structural constraints of the PrP 27-30 crystals, positioning residues 141-176 and the N-linked sugars appropriately. Our parallel left-handed beta-helical model provides a coherent framework that is consistent with many structural, biochemical, immunological, and propagation features of prions. Moreover, the parallel left-handed beta-helical model for PrP(Sc) may provide important clues to the structure of filaments found in some other neurodegenerative diseases.
Figures



References
-
- Prusiner, S. B., Scott, M. R., DeArmond, S. J. & Cohen, F. E. (1998) Cell 93, 337-348. - PubMed
-
- Kelly, J. W. (1998) Curr. Opin. Struct. Biol. 8, 101-106. - PubMed
-
- Sunde, M., Serpell, L. C., Bartlam, M., Fraser, P. E., Pepys, M. B. & Blake, C. C. (1997) J. Mol. Biol. 273, 729-739. - PubMed
-
- Sunde, M. & Blake, C. C. F. (1998) Q. Rev. Biophys. 31, 1-39. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

