Intramembrane proteolysis and endoplasmic reticulum retention of hepatitis C virus core protein
- PMID: 15163730
- PMCID: PMC416534
- DOI: 10.1128/JVI.78.12.6370-6380.2004
Intramembrane proteolysis and endoplasmic reticulum retention of hepatitis C virus core protein
Abstract
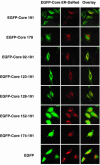
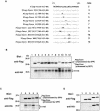
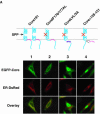
Hepatitis C virus (HCV) core protein is suggested to localize to the endoplasmic reticulum (ER) through a C-terminal hydrophobic region that acts as a membrane anchor for core protein and as a signal sequence for E1 protein. The signal sequence of core protein is further processed by signal peptide peptidase (SPP). We examined the regions of core protein responsible for ER retention and processing by SPP. Analysis of the intracellular localization of deletion mutants of HCV core protein revealed that not only the C-terminal signal-anchor sequence but also an upstream hydrophobic region from amino acid 128 to 151 is required for ER retention of core protein. Precise mutation analyses indicated that replacement of Leu(139), Val(140), and Leu(144) of core protein by Ala inhibited processing by SPP, but cleavage at the core-E1 junction by signal peptidase was maintained. Additionally, the processed E1 protein was translocated into the ER and glycosylated with high-mannose oligosaccharides. Core protein derived from the mutants was translocated into the nucleus in spite of the presence of the unprocessed C-terminal signal-anchor sequence. Although the direct association of core protein with a wild-type SPP was not observed, expression of a loss-of-function SPP mutant inhibited cleavage of the signal sequence by SPP and coimmunoprecipitation with unprocessed core protein. These results indicate that Leu(139), Val(140), and Leu(144) in core protein play crucial roles in the ER retention and SPP cleavage of HCV core protein.
Figures






References
-
- Aizaki, H., Y. Aoki, T. Harada, K. Ishii, T. Suzuki, S. Nagamori, G. Toda, Y. Matsuura, and T. Miyamura. 1998. Full-length complementary DNA of hepatitis C virus genome from an infectious blood sample. Hepatology 27:621-627. - PubMed
-
- Chang, S. C., J. H. Yen, H. Y. Kang, M. H. Jang, and M. F. Chang. 1994. Nuclear localization signals in the core protein of hepatitis C virus. Biochem. Biophys. Res. Commun. 205:1284-1290. - PubMed
-
- Choo, Q. L., G. Kuo, A. J. Weiner, L. R. Overby, D. W. Bradley, and M. Houghton. 1989. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science 244:359-362. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

